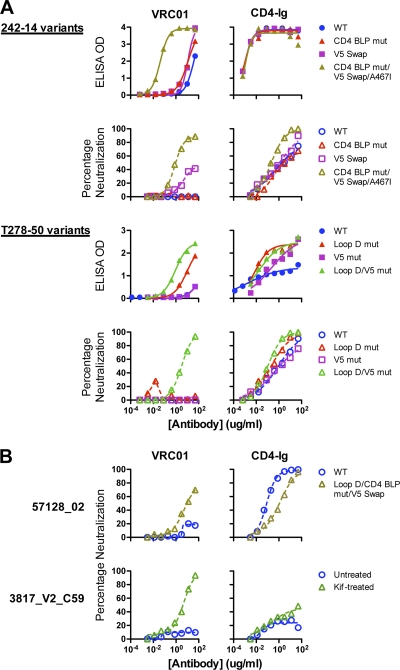
Fig. 4.
Binding and neutralization of reversion mutants and chimeras of VRC01-resistant viruses. (A) VRC01-resistant viruses 242-14 and T278-50 and their reversion mutants. Curves show binding and neutralization with VRC01 or CD4-Ig. The specific reversion mutants and chimeras are described in Table 2. (B) Analysis of two Env pseudoviruses that were resistant to VRC01 despite recognition of the viral gp120. gp120 of virus 57128_02 bound to VRC01 with an EC50 of 8.7 μg/ml (Table 1) yet was resistant to VRC01 neutralization. With additional reversion mutations in loop D, CD4 binding loop, and V5, the mutant virus was moderately neutralized by VRC01 (top). Virus 3817_V2_C59 was resistant to VRC01 neutralization (bottom) despite strong recognition of its gp120 by VRC01 (EC50 of 0.04 μg/ml, Table 1). When the virus was produced in the presence of the glycan modification inhibitor kifunensin, it became sensitive to VRC01-mediated neutralization (bottom).