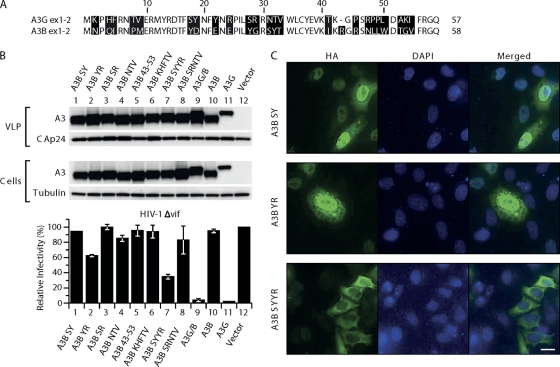
Fig. 3.
Mutational analysis of the first 60 amino acids in A3B. (A) Amino acids in A3B were replaced with residues present in A3G at corresponding positions. 293T cells were cotransfected with the A3B mutants with pCMVHIV-1Δvif, pUCHR-GFPLuc, and pCMV-VSV-G; HeLaP4 cells were infected with the resultant VLPs, and luciferase activity was measured at 48 h postinfection. The infectivity of virus produced in the presence of the different mutants relative to that in the absence of A3 proteins, set at 100%, is shown. Data are the means from 3 independent experiments, and error bars represent the standard deviations. (B) Expression of A3 mutants in cells and packaging into the VLPs. The presence of A3 proteins in virus and cell extracts was analyzed by immunoblotting with anti-HA antibody probing. Tubulin and HIV-1 CA proteins were also detected to verify equal cell and virion loading. (C) Localization of A3B mutants. HeLaP4 cells were transiently transfected with plasmids encoding the mutants A3B SY (Y18D, S19Y) A3B YR (E22Y, Y24R), and A3B SYYR (Y18D, S19Y, E22Y, Y24R). Cells were fixed, permeabilized, and stained with mouse anti-HA antibodies and Alexa Fluor 488 goat anti-mouse IgG. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).