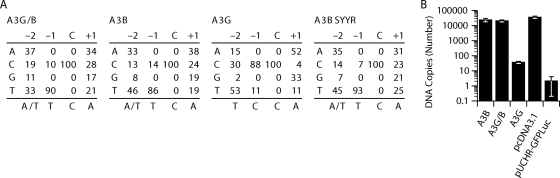
Fig. 5.
Comparison of the sequence contexts preferred by A3B, A3G, and chimera A3G/B. (A) The matrices show the frequency (in percent) of the four bases at positions −2, −1, and + 1 relative to the dC residue, which was deaminated. The consensus sequences are shown on the bottom of each matrix. (B) Amounts of late RT products quantified by the U5Ψ primer-probe set. Viral DNA levels were determined at 24 h after infection with HIV-1Δvif virus produced in the presence of the indicated A3 proteins or pcDNA3.1. To determine DNA contamination, pUCHR-GFPLuc vector was used. Error bars in the graph represent the standard deviations from the means from three independent experiments.