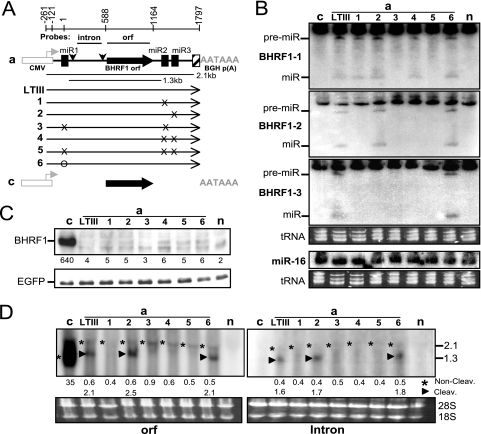
Fig. 3.
Blocking Drosha cleavage of the exon does not affect the mRNA level and encoded protein expression. The 293 cells were cotransfected by an EGFP-expressing plasmid and the wild-type or mutant BHRF1 construct. At 24 h posttransfection, the levels of BHRF1 protein or miRNA and mRNA were determined by Western or Northern analysis. The expression of EGFP was examined as an internal control. (A) Diagram of BHRF1 constructs. Shown are the CMV promoter (open box) and its transcription orientation (horizontal arrowhead), BGH polyadenylation signal (AATAAA), authentic BHRF1 polyadenylation signal (hatched box), ORF and intron probes, BHRF1 ORF (filled arrow), pre-miR-BHRF1-1 (miR1), pre-miR-BHRF1-2 (miR2), pre-miR-BHRF1-3 (miR3), splice donor and acceptor (vertical arrowheads), 2.1- and 1.3-kb BHRF1 RNA transcripts, and mutations (× or O). The BHRF1 constructs are named on the left. (B) Northern analysis of the pre-miRNA (pre-miR) and mature miRNA (miR). n, mock transfection. (C) Western analysis of protein expression. Numbers on the bottom indicate the relative intensity of specific BHRF1 protein bands. n, mock transfection. (D) Northern analysis of the BHRF1 RNA transcripts by using 32P-labeled ORF or intron probe. The positions of 2.1- and 1.3-kb RNA bands are indicated on the right in kilobases. The numbers on the bottom indicate the relative intensities of noncleavage (*) or Drosha cleavage (▶) BHRF1 RNA bands. 18S, 18S rRNA; 28S, 28S rRNA; n, mock transfection.