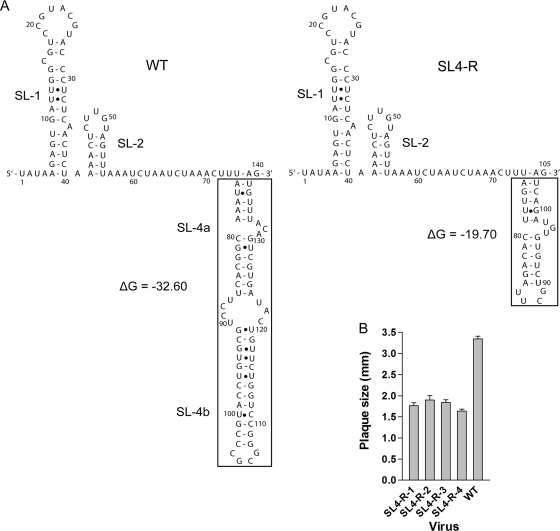
Fig. 7.
Secondary structure and average plaque size of the SL4-R mutant. (A, left) The predicted secondary structures of the WT 5′ 140 nt (positions 1 through 140) including SL1, SL2, and SL4 in our model (14, 24). ΔG = −32.60 kcal/mol. (A, right) The secondary structures of SL4-R mutant (positions 1 through 105) predicted by Mfold 3.0, including SL1, SL2, and SL4, replaced by a shorter sequence-unrelated stem-loop. ΔG = −19.70 kcal/mol. The structures in the boxes represent the corresponding replacement. (B) Average plaque sizes of the four plaque-purified isolates of the SL4-R mutant.