Figure 4.
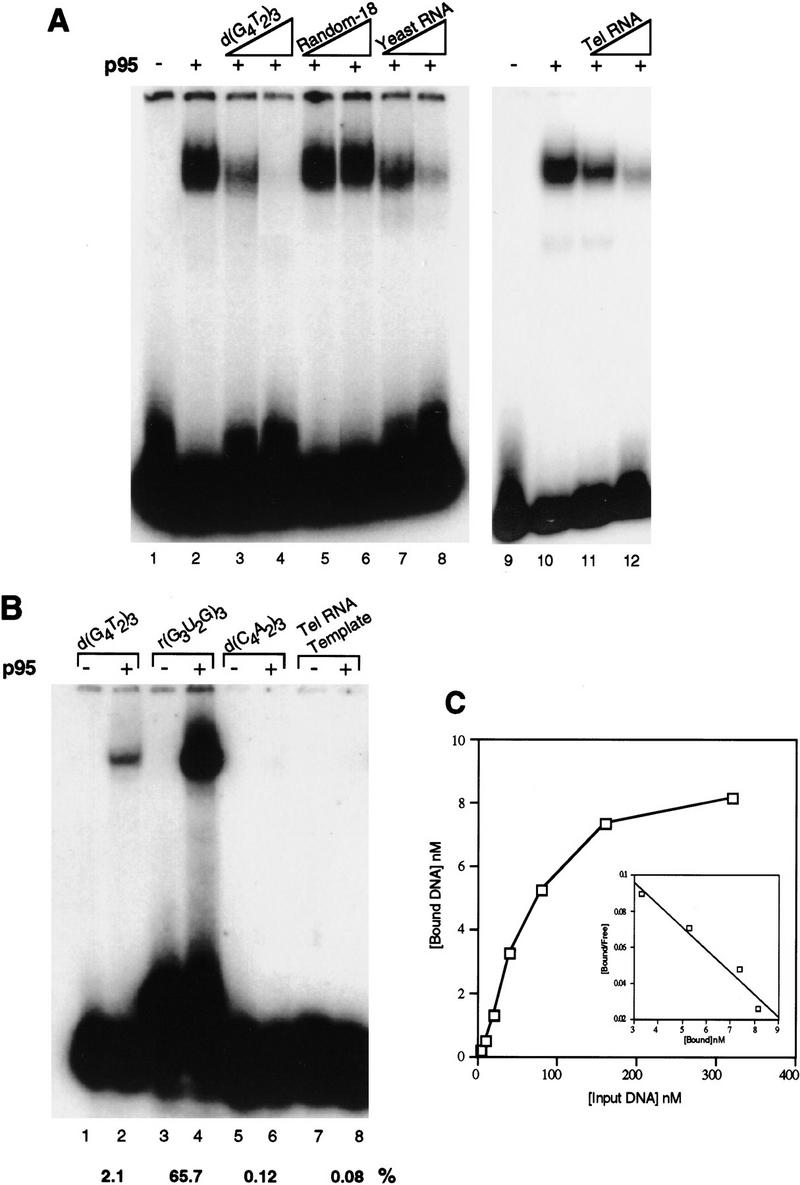
Single-stranded, G-rich telomeric sequence is preferred for interaction with p95. (A) Mobility shift assay with p95 at 100 nm and radiolabeled d(G4T2)3 at 20 nm (double-stranded DNA competitor poly[d(I–C)], only; see Materials and Methods). (Lanes 1,9) probe alone; (lanes 2,10) mobility shift induced by p95. Lanes 3–6 and 11 and 12 have the indicated unlabeled competitor DNA or RNA added at 50- or 500-fold the molar concentration of probe: (lanes 3,4) d(G4T2)3; (lanes 5,6) a random 18-nucleotide DNA sequence (Random-18); (lanes 11,12) T. thermophila telomerase RNA. Because molar equivalents could not be determined for total yeast RNA, lanes 7 and 8 have total unlabeled yeast RNA added at 100 ng and 1 μg [roughly double the mass of d(G4T2)3 in (lanes 3 and 4)]. The p95 used in lanes 7 and 8 was a side fraction; the weak, lower mobility shift band in these lanes likely represents binding to a breakdown product of p95 in this fraction. (B) Direct mobility shift assays of radiolabeled telomeric and nontelomeric DNA and RNA sequences by p95 (double-stranded DNA competitor poly[d(I–C)], only). Probes were used at 20 nm with 100 nm p95 as follows: (lanes 1,2) d(G4T2)3; (lanes 3,4) r(G3U2G)3; (lanes 5,6) d(C4A2)3; (lanes 7,8) the telomerase RNA template region, rCAACCCCAAAAAUCUAGUGC. The percentage probe bound in each case was quantitated and indicated below the appropriate lane as %. (C) The binding of p95 to single-stranded, G-rich telomeric DNA was measured using 25 nm protein and twofold increases of d(G4T2)3 from 5 to 320 nm. A Scatchard plot (inset) was derived from data at the four highest protein concentrations (40–320 nm).
