Abstract
Inoculation of soybean (Glycine max) plants with Phakopsora pachyrhizi, the causal organism of Asian soybean rust, elicits a biphasic response characterized by a burst of differential gene expression in the first 12 h. A quiescent period occurs from 24 to 48 h after inoculation, in which P. pachyrhizi continues to develop but does not elicit strong host responses, followed by a second phase of intense gene expression. To correlate soybean responses with P. pachyrhizi growth and development, we inoculated the soybean cultivar Ankur (accession PI462312), which carries the Rpp3 resistance gene, with avirulent and virulent isolates of P. pachyrhizi. The avirulent isolate Hawaii 94-1 elicits hypersensitive cell death that limits fungal growth on Ankur and results in an incompatible response, while the virulent isolate Taiwan 80-2 grows extensively, sporulates profusely, and produces a compatible reaction. Inoculated leaves were collected over a 288-h time course for microarray analysis of soybean gene expression and microscopic analysis of P. pachyrhizi growth and development. The first burst in gene expression correlated with appressorium formation and penetration of epidermal cells, while the second burst of gene expression changes followed the onset of haustoria formation in both compatible and incompatible interactions. The proliferation of haustoria coincided with the inhibition of P. pachyrhizi growth in the incompatible interaction or the beginning of accelerated growth in the compatible interaction. The temporal relationships between P. pachyrhizi growth and host responses provide an important context in which to view interacting gene networks that mediate the outcomes of their interactions.
Interactions between plants and pathogens generally fall under the categories of compatible or incompatible. In compatible interactions, a host plant supports the development and spread of a pathogen, frequently resulting in disease. In incompatible interactions, the plant is resistant to the growth of a pathogen, resulting in no or reduced disease. Plant defense responses are triggered by both nonspecific and specific recognition of potential pathogens. One type of defense, mediated by nonspecific recognition, is termed pathogen-associated molecular pattern-triggered immunity (PTI) and is triggered by conserved properties of a particular class of microbes (Jones and Dangl, 2006; Mackey and McFall, 2006; Bent and Mackey, 2007). For example, plant cells recognize certain bacteria through the perception of fragments of the highly conserved flagellin protein (Felix et al., 1999; Gómez-Gómez et al., 1999; Meindl et al., 2000), while many fungi are detected through wall fragments derived from the chitin polymer (Wan et al., 2008a, 2008b). Successful pathogens are able to overcome PTI through the action of effector proteins that actively suppress host defense (Jones and Dangl, 2006; Mackey and McFall, 2006; Bent and Mackey, 2007). In turn, plants have evolved to harbor resistance (R) genes that mediate specific recognition of effectors produced by particular races or species of pathogens, thus triggering massive defense responses (Jones and Dangl, 2006; Mackey and McFall, 2006; Bent and Mackey, 2007). This effector-triggered immunity (ETI) frequently results in a hypersensitive response that curtails pathogen invasion.
Biotrophic pathogens such as the rust fungi (order Uredinales) require living host tissues in order to complete their life cycles. Like other pathogens, rusts must escape or avoid PTI as they progress through distinct phases of their life cycle, and they elicit ETI through the secretion of effectors (Ravensdale et al., 2011). Some R genes mediate early and rapid responses that arrest pathogen growth with no macroscopic symptoms. Other R genes allow growth for a short time until pathogen recognition occurs, at which time a macroscopic hypersensitive response develops. Such responses may completely halt pathogen growth, or the pathogen may continue to grow only at severely reduced rates. The type of defense response exhibited by the host plant is influenced by the timing of effector delivery and effector interactions with cognate R proteins. The timing of interactions between rusts and their hosts is strongly influenced by the pathogen’s formation of specialized feeding cells known as haustoria (Voegele and Mendgen, 2003). The importance of rust haustoria in the production of effectors was recently demonstrated for flax (Linum usitatissimum) rust. Flax rust haustoria produce effector proteins that are secreted and taken up by flax cells, eliciting ETI (Catanzariti et al., 2006; Dodds et al., 2006; Ellis et al., 2007a; Rafiqi et al., 2010).
Phakopsora pachyrhizi is an obligate, biotrophic, plant-pathogenic fungus that causes the disease commonly known as Asian soybean rust (ASR). P. pachyrhizi primarily colonizes leaf tissue, and under favorable environmental conditions, infection can result in yield losses ranging from 10% during mild disease pressure to 80% during severe epidemics (Ogle et al., 1979; Bromfield, 1984; Patil et al., 1997). Six soybean (Glycine max) loci confer either immunity (Rpp1) or resistance with little or no sporulation (Rpp1b, Rpp2, Rpp3, Rpp4, and Rpp5; Cheng and Chan, 1968; Hidayat and Somaatmadja, 1977; Singh and Thapliyal, 1977; Bromfield and Hartwig, 1980; McLean and Byth, 1980; Hartwig and Bromfield, 1983; Hartwig, 1986; Monteros et al., 2007; Garcia et al., 2008). The destructive potential of P. pachyrhizi coupled with its recent introduction into the major soybean-growing countries of the Western Hemisphere has generated interest in understanding the molecular interactions of P. pachyrhizi with its soybean host and with nonhost plants (Loehrer et al., 2008; Hoefle et al., 2009; Goellner et al., 2010).
mRNA transcript profiling of compatible and incompatible soybean responses to P. pachyrhizi demonstrated that soybean plants with or without the Rpp2 resistance gene respond strongly to P. pachyrhizi infection within the first 12 h after inoculation (hai; van de Mortel et al., 2007). This initial response was followed by an interval of time in which the fungus grew virtually undetected and unchallenged. A second response to P. pachyrhizi occurred in both resistant and susceptible plants but happened earlier and with a greater magnitude in resistant plants, suggesting that ETI was elicited at this time. The striking biphasic response suggested that different stages of fungal development were associated with the plant responses. Previous studies of P. pachyrhizi growth and development provided an estimate of fungal growth stages over time (Marchetti et al., 1975; Bonde et al., 1976; Deverall et al., 1977; McLean, 1979; Koch et al., 1983), but correlations between soybean gene expression responses and P. pachyrhizi development have not been reported.
To establish relationships between P. pachyrhizi growth stages and soybean responses, and to determine whether the biphasic response occurs in other P. pachyrhizi-soybean interactions, we devised an experimental system that minimized genomic effects caused by different soybean genotypes. We infected the soybean cv Ankur (accession PI462312) carrying the Rpp3 resistance gene with incompatible (Hawaii 94-1 [HW94-1]) and compatible (Taiwan 80-2 [TW80-2]) isolates of P. pachyrhizi (Bonde et al., 2006; Hyten et al., 2009). mRNA transcript profiling and microscopic analyses were performed on plants sampled over a time course spanning early, middle, and late stages of infection. Using a single soybean genotype increased our ability to detect small yet significant changes in gene expression and allowed us to attribute detected gene expression changes specifically to differences between the two P. pachyrhizi isolates. The results demonstrated that the early gene expression responses in the compatible and incompatible interactions were correlated with the penetration of epidermal cells at infection sites. The second response was characterized by more rapid and extensive alterations in gene expression in response to the avirulent isolate, and it correlated well with the proliferation of haustoria.
RESULTS
Experimental Design and Verification of P. pachyrhizi Infection
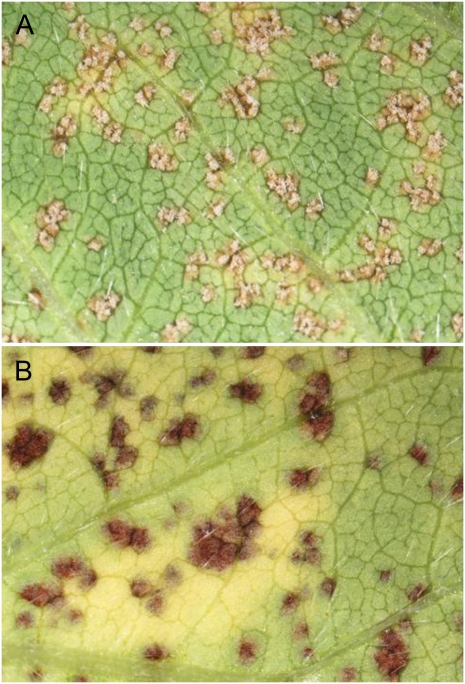
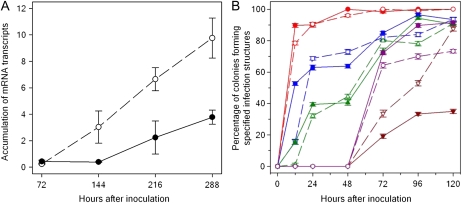
Ankur (Rpp3; PI462312) plants were inoculated with urediniospores from the avirulent P. pachyrhizi isolate HW94-1 or from the virulent isolate TW80-2. Leaf samples were collected for microscopy at 12, 24, 48, 72, 96, 120, 144, 216, and 288 hai and for mRNA profiling at 12, 24, 72, 144, 216, and 288 hai. Successful inoculation was verified at 288 hai (12 d) by the appearance of reddish-brown (RB) lesions on the abaxial side of HW94-1-inoculated leaves (Fig. 1A) and similar numbers of tan lesions on TW80-2-inoculated leaves (Fig. 1B). These data thus confirmed the expected phenotypes for the incompatible and compatible interactions. RNA was extracted from leaves that were inoculated with HW94-1, TW80-2, or a mock-inoculation solution that did not contain spores. Three biological replicates were performed with samples randomized for treatment and collection time within each replicate (block). Resistant and susceptible phenotypes were verified by measuring the abundance of a constitutively expressed P. pachyrhizi α-tubulin mRNA in each RNA sample (Fig. 2A). Quantitative reverse transcription (qRT)-PCR assays demonstrated that similar quantities of P. pachyrhizi α-tubulin transcripts were present in HW94-1- and TW80-2-infected soybean leaves through 72 hai. Starting at 144 hai, the P. pachyrhizi α-tubulin transcript abundance dramatically increased in the compatible interaction, indicating prolific fungal growth and colonization. In contrast, the P. pachyrhizi α-tubulin transcript increased relatively slowly after 144 hai in the incompatible interaction, confirming that the expected resistance response had occurred. These results demonstrated that P. pachyrhizi infection was successful in this experiment and that the selected P. pachyrhizi isolates had the expected infection phenotypes in Rpp3 plants.
Figure 1.
P. pachyrhizi lesions 288 hai (12 d after inoculation) on the adaxial leaf surface of Ankur (PI462312) soybean plants carrying the Rpp3 resistance gene. A, Tan-colored, susceptible-type lesions with abundant uredinia after infection with the virulent P. pachyrhizi isolate TW80-2. B, RB, resistant-type lesions caused by the avirulent P. pachyrhizi isolate HW94-1.
Figure 2.
Quantitative observations of P. pachyrhizi infection kinetics on Ankur (PI462312) soybean plants during the incompatible (isolate HW94-1; black symbols and solid lines) and compatible (isolate TW80-2; white symbols and dotted lines) interactions. A, Differential accumulation of P. pachyrhizi α-tubulin mRNA relative to soybean ubiquitin-3 in soybean leaves determined by qRT-PCR (mean ± se; n = 3). B, Microscopic observations of infection structures in single infection colonies: formation of appressoria (red), infection hyphae (blue), intercellular hyphae (green), haustoria (brown), and discoloration of host cells (purple).
Microscopic Observation of P. pachyrhizi Infection
To better correlate fungal growth and development with soybean gene expression, we monitored P. pachyrhizi growth microscopically at each of the six time points used for expression profiling and also included 48-, 96-, and 120-hai samples to allow analyses of fungal development at an even higher resolution.
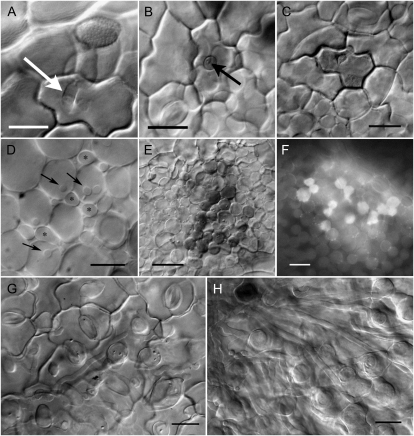
Urediniospores produced a single germ tube that ranged from just a few micrometers to over 300 μm in length (data not shown). Most germinating urediniospores had produced appressoria by 12 hai (Fig. 2B). The appressoria formed directly over epidermal cells or at the anticlinal wall junction between adjacent epidermal cells. Penetration was direct, and infection hyphae were produced within the epidermal cells (Fig. 3A). Infection hyphae were present at a significant proportion of infection sites by 12 hai and were present at most infection sites by 24 hai (Fig. 2B). More HW94-1 urediniospores had penetrated and produced infection hyphae at 12 hai than TW80-2, demonstrating that HW94-1 developed slightly faster during these early infection processes. By 24 hai, penetration and the development of infection hyphae were similar for the two isolates (Fig. 2B), and many of the infection hyphae had emerged from the infected epidermal cell to establish intercellular hyphae within the palisade mesophyll (Fig. 3B). The infected epidermal cells became noticeably discolored by 72 to 96 hai after infection with either P. pachyrhizi isolate (Fig. 3C).
Figure 3.
Micrographs of P. pachyrhizi infection structures on Ankur (PI462312) soybean plants carrying the Rpp3 resistance gene. A, Infection hypha (arrow) in epidermal cell 24 hai with avirulent P. pachyrhizi isolate HW94-1. Urediniospore and appressorium can be seen slightly out of focus. B, Infection hypha within an epidermal cell 24 hai with virulent P. pachyrhizi isolate TW80-2. The arrow indicates a penetration peg leading to an intercellular hypha in the palisade mesophyll below. C, Discolored infected epidermal cell 96 hai with isolate HW94-1. D, Haustoria (arrows) and intercellular hyphae (asterisks) in palisade mesophyll 96 hai with isolate TW80-2. E, Discolored palisade mesophyll cells in leaf infected with isolate HW94-1 at 144 hai. F, Autofluorescence of discolored palisade mesophyll cells at 120 hai with isolate HW94-1. Deposits that accumulate in the wall region between mesophyll cells are also highly autofluorescent. G, Sparse hyphae of isolate HW94-1 in spongy mesophyll tissue at 144 hai. H, Abundant hyphae of isolate TW80-2 in spongy mesophyll tissue, radiating out from the center of an infection site at 144 hai. Bars = 20 μm except for D (10 μm), E (40 μm), and F (30 μm).
However, differences between the compatible and incompatible interactions became apparent between 72 and 96 hai. At these time points, colonies of the virulent isolate (TW80-2) consisted of abundant intercellular hyphae, and haustoria were seen in many of the palisade mesophyll cells (Fig. 3D). Fewer haustoria were present in colonies of the avirulent HW94-1 isolate (Fig. 2B). Also during this time period, discoloration was observed in the colonized palisade mesophyll tissues (Fig. 3E), and more discolored cells were observed in tissues colonized by HW94-1 than in tissues colonized by TW80-2 (Fig. 2B). These differences in haustoria frequency and cell discoloration became even greater by 120 hai (Fig. 2B). The discolored cells exhibited bright autofluorescence, as did the deposits of wall material that were often seen between palisade mesophyll cells in tissues colonized by the avirulent HW94-1 isolate (Fig. 3F). The colonies of TW80-2 expanded much more rapidly than HW94-1 colonies after 96 hai. This difference in growth rate became striking at 144 hai, when HW94-1 (Fig. 3G) and TW80-2 (Fig. 3H) were colonizing the spongy mesophyll tissues.
Expression of Soybean Genes in Response to Compatible and Incompatible Interactions with P. pachyrhizi Isolates
The abundance of soybean mRNA transcripts was assayed using the Affymetrix GeneChip Soybean Genome Array, and these data were deposited in the Gene Expression Omnibus database (accession no. GSE29740; Edgar et al., 2002) and the Plant Expression database (accession no. GM36; Wise et al., 2007). The Ankur (Rpp3) genetic background is the same for all the soybean-P. pachyrhizi interactions, allowing us to make direct comparisons between infected and mock-inoculated plants as well as between incompatible and compatible interactions. Statistical inference was conducted using a separate general linear model analysis for each probe set. Significantly differentially expressed probe sets were identified by statistical analyses of the data from the six different time points. To make the data set a manageable size, we filtered the probe sets using a 0.01% false discovery rate (FDR; q ≤ 0.0001, derived from values of P < 5.15 × 10−5; Supplemental Table S1; see “Materials and Methods”; Nettleton, 2006). At the 0.01% FDR, probe sets were identified whose pattern of expression over the time course in mock-inoculated plants differed from the pattern of expression in the HW94-1- and/or the TW80-2 treated plants. Of these, 1,799 probe sets were differentially expressed in response to both HW94-1 and TW80-2, 27 were unique to the TW80-2 treatment, and 6,647 were unique to the HW94-1 treatment. For simplicity, we hereafter refer to these probe sets as differentially expressed genes (DEGs).
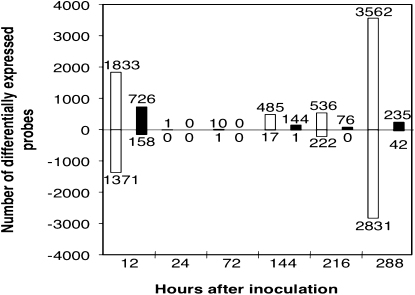
We also conducted statistical analyses at individual time points for the HW94-1 versus mock-inoculated and for the TW80-2 versus mock-inoculated comparisons using a 0.01% FDR cutoff. The number of DEGs were plotted at each time point (Fig. 4), illustrating a biphasic pattern of differential gene expression for both induced and repressed genes, similar to our previous analyses involving a different fungal isolate and two different soybean genotypes (van de Mortel et al., 2007). In the HW94-1 treatment, there were 1,833 up-regulated and 1,371 down-regulated DEGs relative to the mock inoculation at 12 hai. However, only a single DEG was identified at the 0.01% FDR level at 24 hai. Differential expression of both up- and down-regulated DEGs increased from 72 hai onward, reaching a maximum at 288 hai (3,562 up-regulated and 2,831 down-regulated). In the TW80-2 treatment, there were 726 up-regulated and 158 down-regulated DEGs at 12 hai, and no differential gene expression was detected at 24 or 72 hai at 0.01% FDR (Fig. 4). Differential gene expression fluctuated after 144 hai, with 145, 76, and 277 DEGs at 144, 216, and 288 hai, respectively. Unlike the incompatible interaction, the most DEGs were observed at 12 hai, and very few genes were down-regulated over the experimental time course. These findings suggest that P. pachyrhizi does not elicit host responses in both compatible and incompatible interactions from 24 to 72 hai. However, beginning at 72 hai, differential gene expression was strongly elicited in the incompatible interaction.
Figure 4.
Number of differentially expressed soybean genes per time point reveals distinct biphasic responses in the incompatible (white bars) and compatible (black bars) P. pachyrhizi-soybean interactions. The values above and below zero represent the numbers of up- and down-regulated genes, respectively, at each time point. These numbers were determined for each time point by identifying genes with q ≤ 0.0001 (0.01% FDR) when treatment with the avirulent P. pachyrhizi isolate HW94-1 was compared with the mock inoculation or when treatment with the virulent P. pachyrhizi isolate TW80-2 was compared with mock inoculation. Values were derived from Supplemental Table S1.
Functional Annotation of P. pachyrhizi-Responsive Genes
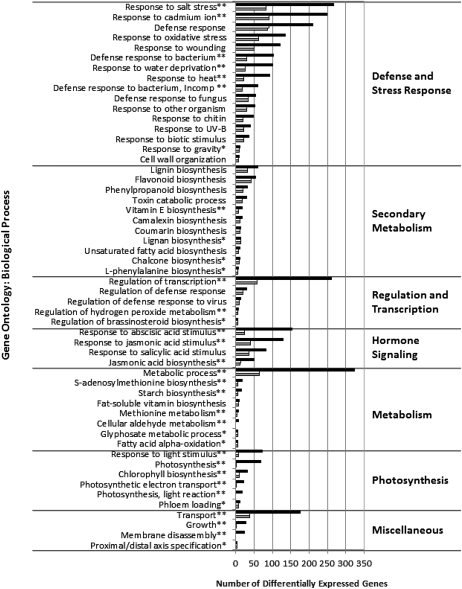
Gene descriptions and functional annotations were downloaded from the SoyBase GeneChip Web site (http://www.soybase.org/AffyChip; “see Materials and Methods”) for the 8,473 DEGs identified in our initial statistical analysis. Functional information included probe set matches to homologous Arabidopsis (Arabidopsis thaliana) genes and their associated Gene Ontology (GO) biological process terms (E ≤ 104; Ashburner et al., 2000). Fisher’s exact test was used to identify GO biological process categories that were significantly overrepresented in each data set (Fig. 5; Supplemental Table S2; Fisher, 1966; Drǎghici et al., 2003). A Bonferroni correction was used to adjust for multiple testing (Bonferroni, 1935). A P value cutoff of 0.05 after correction was used. This analysis identified 54 GO biological process categories that were overrepresented. These were grouped into seven broad categories: defense and stress responses, secondary metabolism, regulation and transcription, hormone signaling, metabolism, photosynthesis, and miscellaneous.
Figure 5.
Overrepresented GO biological process classification of P. pachyrhizi-responsive genes as determined by Fisher’s exact test (Fisher, 1966). The total numbers of differentially expressed genes in the incompatible and compatible P. pachyrhizi-soybean interactions are represented by black and white bars, respectively. DEGs common to both interactions are represented by gray bars. Since a DEG could be associated with multiple GO processes, it could be represented multiple times. Individual GO categories have been grouped into broader functional categories, which are labeled. Only significantly (P ≤ 0.05) overrepresented GO categories are shown. One asterisk following a GO description indicates that the GO category is overrepresented only in the compatible interaction. Two asterisks following the GO description indicate that the GO category is overrepresented only in the incompatible interaction. For GO category identifiers and the complete data set, see Supplemental Table S2.
Fourteen GO categories were significant only in the incompatible interaction (Fig. 5; Supplemental Table S2). In this case, differences in the numbers of incompatible DEGs versus the compatible DEGs in these categories ranged from 1.4- to 34.5-fold. In three categories, photosynthesis (light reaction), cellular aldehyde metabolic process, and membrane disassembly, no DEGs were identified in the compatible interaction. Twenty GO functional classes were overrepresented in both the incompatible and compatible interactions, including defense and stress response (six of 16 GO classes in this subcategory), secondary metabolism (seven of 11 GO classes), regulation and transcription (two of five GO classes), hormone signaling (one of five GO classes), and metabolism (one of seven GO classes). In each of these cases, while both interactions were significant, far more DEGs were indicated in the incompatible interaction (Fig. 5; Supplemental Table S2).
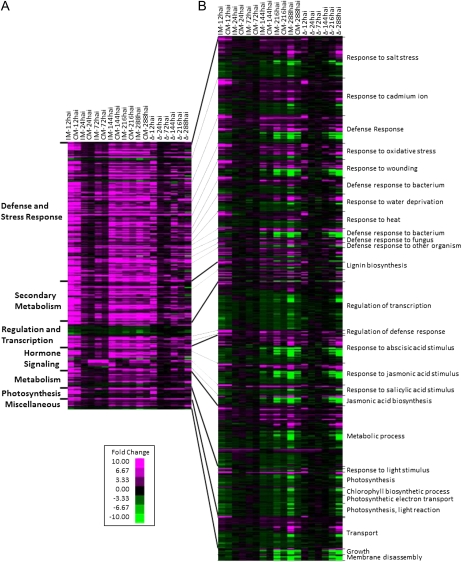
To better understand the differences between compatible and incompatible interactions, we compared expression in the 1,799 DEGs common to the incompatible and compatible interactions (Fig. 6A). To put the DEGs in a biological context, only the 616 genes assigned to an overrepresented GO category identified in Figure 5 were examined. Hierarchical clustering of expression data was performed within each GO category to cluster DEGs with similar expression patterns (Fig. 6A). In general, both interactions were quite similar. For almost all GO categories, a biphasic induction of gene expression was observed. At 12 hai, the majority of common genes were up-regulated in both the compatible and incompatible interactions. Gene expression returned to mock-inoculated levels at 24 hai, which was followed by a second round of differential expression that began at 72 hai in the incompatible interaction, while it was not detected in the compatible interaction until 144 hai. Only the GO category regulation of transcription deviated from this pattern. Within this category, the expression of a majority of genes was down-regulated relative to mock inoculation. While a slight biphasic response was seen, expression differences between mock-inoculated and infected tissues were much smaller.
Figure 6.
Changes in soybean gene expression demonstrate a biphasic response to P. pachyrhizi infection. To put genes in a biological context, only DEGs mapping to an overrepresented GO biological process category (Fig. 5) are shown. Data are shown for all 54 overrepresented GO categories, but only GO categories containing more than 25 DEGs are labeled. Within each GO category, hierarchical clustering was used to group DEGs with similar expression patterns. A, The seven major GO categories are labeled at left. The first 12 columns (IM-12hai to CM-288hai) contain gene expression analyses of the 1,799 DEGs common to the incompatible and compatible P. pachyrhizi-soybean interactions compared with mock-inoculated plants (IM and CM, respectively) over six time points. Black boxes represent no change in gene expression compared with the uninfected mock-inoculated treatment, magenta indicates up-regulation, and green indicates down-regulation. More intense colors represent greater fold change (log2 transformed), as shown on the scale. The final six columns (Δ-12hai to Δ-288hai) directly compare fold change in gene expression for each time point between the incompatible versus mock-inoculated and compatible versus mock-inoculated interactions. Black signifies equal expression in the incompatible and compatible interactions, magenta signifies greater expression in the incompatible interaction, while green signifies greater expression in the compatible interaction. B, Gene expression analyses of the 6,647 DEGs unique to the incompatible interaction. The labels to the right indicate GO subcategories of the seven major categories listed in A. Black indicates no change in expression relative to mock-inoculated plants, while magenta and green indicate up- and down-regulation, respectively. For both panels, a DEG may be represented once per GO category but multiple times across different GO categories.
Our analyses thus far suggested that genes common to the compatible and incompatible interactions were expressed in similar patterns but with different intensities. Since our experiments used a single soybean genotype, we could directly compare the difference in fold change relative to mock inoculation between the two interactions at each time point. Using hierarchical clustering, we saw substantial differences in the compatible and incompatible interactions within the common genes. At 12 hai, the incompatible interaction had significantly greater up-regulation than the compatible interaction. While the dip in gene expression was still seen at 24 and 72 hai, clear signs of greater differential expression in the incompatible interaction were seen from 72 hai onward.
We also used the GO information to examine the 6,647 DEGs unique to the incompatible interaction. Of these 6,647 DEGs, 2,086 DEGs mapped to the statistically overrepresented GO categories in Figure 5. When expression patterns between the common and unique gene sets were examined, the differences were striking (Fig. 6). Unlike the common genes that tended to be up-regulated relative to the mock inoculation, the expression patterns of genes unique to the incompatible interaction were more diverse. For example, in the defense and stress subcategory, approximately half of the 715 unique genes were up-regulated by P. pachyrhizi infection, while the rest were down-regulated. Furthermore, for DEGs slightly down-regulated at 12 hai, we saw increasing down-regulation from 144 to 288 hai. In the broader category of secondary metabolism, an additional 84 DEGs were identified, with equal numbers of DEGs up- and down-regulated. In the regulation and transcription subcategory, an additional 215 unique genes were differentially expressed. While the majority of these genes were down-regulated, those associated with transcriptional regulation of the defense response were up-regulated. In order to understand how gene expression and function are related to plant defense, we will focus below on the GO biological process categories of hormone signaling, regulation and transcription, defense and stress responses, metabolism, and photosynthesis.
Hormone Signaling
Four GO categories involved in hormone signaling were overrepresented in the incompatible interaction: responses to salicylic acid, jasmonic acid, and abscisic acid stimuli as well as jasmonic acid biosynthesis (Fig. 6). Only the response to salicylic acid stimulus category was overrepresented in the compatible interaction. For DEGs common to both interactions (Fig. 6A), most were up-regulated by P. pachyrhizi infection, with greater expression in the incompatible interaction. However, for DEGs unique to the incompatible interaction (Fig. 6B), we saw both up- and down-regulation in response to P. pachyrhizi. All three hormones are involved in defense, wounding, or abiotic stress responses (Davies, 2010). Salicylic acid is involved in defense against biotrophic pathogens and in systemic acquired resistance (Bari and Jones, 2009; Van Verk et al., 2009). In contrast, jasmonic acid, a salicylic acid antagonist, is usually involved in defense against insects and necrotrophic pathogens. Abscisic acid acts as a negative regulator of plant defenses. In this experiment, the incompatible interaction appears to have activated signaling through the salicylic acid, jasmonic acid, and abscisic acid pathways.
Regulation and Transcription
Four GO biological process classes associated with regulation and transcription were overrepresented in the incompatible interaction (Fig. 5). Two were common to both interactions, while regulation of transcription and hydrogen peroxide metabolism were unique to the incompatible interaction. Only 7% of probe sets present on the array in the regulation of transcription category were differentially expressed in the compatible interaction, while 30% were differentially expressed in the incompatible interaction.
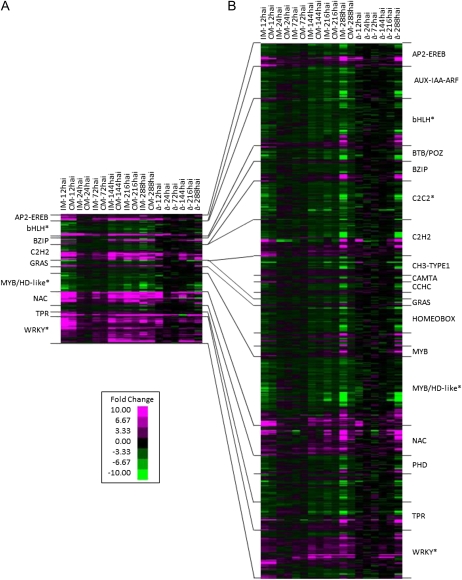
To test if particular transcription factor classes were differentially expressed, we used the soybean locus name assigned by the whole soybean genome assembly (Schmutz et al., 2010) to cross-reference the DEGs with SoyDB, a soybean transcription factor database (Wang et al., 2010). Sixty-three transcription factor classes, representing 2,194 probe sets, were identified on the array. By combining this information with differential gene expression data, we could use Fisher’s exact test (Fisher, 1966; Drǎghici et al., 2003) with a Bonferroni correction (Bonferroni, 1935) to identify overrepresented transcription factor classes in our data. While no transcription factor families were significantly overrepresented in the compatible interaction, WRKY (P = 0), bHLH (P < 0.005), MYB-HD like (P < 0.01), and C2C2 (Zn) CO-like (P < 0.02) transcription factors were overrepresented in the incompatible interaction. Expression patterns of the 118 and 479 differentially expressed transcription factors common to both interactions and unique to the incompatible interaction, respectively, are shown in Figure 7. In this heat map, the transcription factors common to both interactions have stronger expression patterns in the incompatible interaction (Fig. 7A). In addition, while some transcription factors were down-regulated at 24 and 72 hai in the compatible interaction, their expression can still be detected in the incompatible interaction. For the transcription factors uniquely expressed in the incompatible interaction (Fig. 7B), we saw increased DEGs in certain families, including AP2-EREB, AUX-IAA-ARF, bHLH, C2C2, C2H2, homeobox, MYB-HD, NAC, TPR, and WRKY, which have diverse biological roles. bHLH (Zhang et al., 2009), homeobox, and C2H2 (He et al., 2010) transcription factors are generally involved in plant development. AUX-IAA-ARF transcription factors regulate auxin responsiveness. C2C2 transcription factors are involved in meristem identity. Others, such as AP2-EREB, MYB, NAC, and WRKY transcription factors, have roles in defense and abiotic stress response (He et al., 2010). While the MYB transcription factors were largely down-regulated in response to P. pachyrhizi, the NAC and WRKY transcription factors were up-regulated.
Figure 7.
Differential expression of soybean transcription factors in response to P. pachyrhizi infection. The SoyDB (Wang et al., 2010) was used to identify all transcription factors on the Soybean GeneChip Genome Array. Expression data are shown for all differentially expressed transcription factors; however, only large families are labeled. Hierarchical clustering was used to group DEGs with similar expression patterns within a transcription factor family. Coloring and data labels are the same as in Figure 6. A, Gene expression patterns of the 118 transcription factors common to the compatible and incompatible P. pachyrhizi-soybean interactions. B, Gene expression patterns of the 479 transcription factors unique to the incompatible interaction. Asterisks signify transcription factor classes significantly overrepresented only in the incompatible interaction.
Defense and Stress Responses
Nine functional classes related to defense or stress responses were overrepresented in both the incompatible and compatible interactions. For each of these classes, differences between gene numbers in the incompatible interaction versus the compatible interaction ranged from 1.25- to 2.47-fold. Genes included in these GO categories code for peroxidases, lipoxygenases, lipases, phenylpropanoid synthesis genes, resistance gene homologs, pathogenesis-related proteins, and thaumatins. Six GO functional classes were overrepresented only in the incompatible interaction. When the numbers of differentially expressed genes from these classes were compared, the differences ranged from 2.76- to 4.18-fold. The largest fold changes were seen in the response to heat, response to water deprivation, and response to bacteria classes. Within each of these classes, additional members of the gene families mentioned above were identified only in the incompatible reaction. For example, the common genes contained five lipoxygenases, while the genes unique to the incompatible interaction contained an additional 18 lipoxygenases. Other gene classes that were unique to the incompatible interaction included ATP synthases, heat shock proteins, NAM proteins, and tubulins. When averaged across all GO categories in defense and stress response, the incompatible interaction had twice as many DEGs as the compatible interaction, with 46% and 23% (respectively) of the genes represented on the array exhibiting differential expression in response to P. pachyrhizi infection.
Primary and Secondary Metabolism
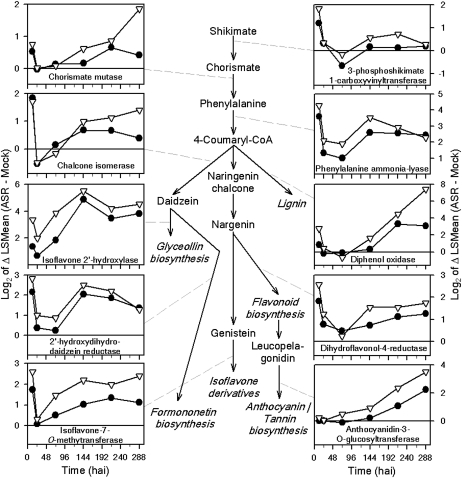
Seven GO biological process classes involved in secondary metabolism were overrepresented in both the incompatible and compatible interactions. Surprisingly, there was little variation in gene expression between the interactions, with differences ranging from 1.17- to 1.93-fold. On average, across all GO categories in secondary metabolism, 62% and 44% of the genes represented on the array were differentially expressed in the incompatible and compatible responses, respectively. We examined expression profiles of genes involved in phenylpropanoid and flavonoid biosynthetic pathways that are known to be involved in plant defense responses through the production of various phytoalexins and cell wall-reinforcing metabolites (Boerjan et al., 2003; La Camera et al., 2004). In general, the flavonoid synthesis genes displayed increased expression at 12 hai (Fig. 8). By 24 hai, however, most genes had returned to basal expression levels and showed no difference between the mock-inoculated and P. pachyrhizi-inoculated samples (Fig. 6). Differential expression was again evident starting at 72 hai, but with different kinetics in the incompatible and compatible interactions. The differential expression of flavonoid genes was generally higher and earlier in the incompatible interaction, and the accumulation of flavonoid mRNAs was greater in the incompatible interaction (Fig. 6).
Figure 8.
Expression profiles of selected genes in the phenylpropanoid biosynthetic pathway in the incompatible (white triangles) and compatible (black circles) P. pachyrhizi-soybean interactions. P. pachyrhizi infection leads to up-regulation of genes in the major flavonoid pathways, as highlighted by the accompanying differential expression profiles of selected genes whose encoded enzymes catalyze reactions in these secondary metabolite pathways.
In primary metabolism, six different GO classes were identified as significantly overrepresented in the incompatible interaction. Only one of these, fat-soluble vitamin biosynthesis, was also overrepresented in the compatible interaction. Of the six GO categories, only metabolic process was represented by more than 10 DEGs. Unlike secondary metabolism, in which compatible and incompatible gene numbers were quite similar, almost five times more genes in the primary metabolism category were differentially expressed in the incompatible interaction.
Photosynthesis Related
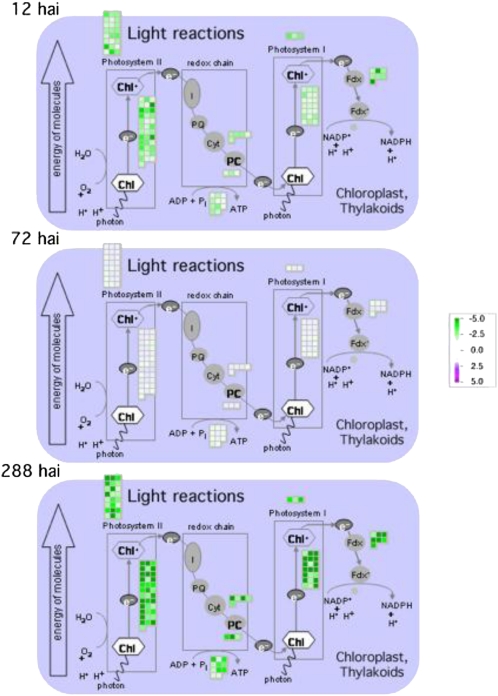
Five statistically overrepresented GO classes in the soybean-P. pachyrhizi data set contained genes primarily related to photosynthesis. Unlike the other general categories described thus far, large differences were observed between the incompatible and compatible interactions. Twenty DEGs were common to both interactions, while 207 were unique to the incompatible interaction. Between 34% and 77% of genes on the array associated with photosynthesis, photosynthetic electron transport, chlorophyll biosynthesis, light reaction, or light response were differentially expressed only in the incompatible interaction. Expression patterns of these photosynthesis-related genes revealed a biphasic pattern of down-regulation, as depicted in Figure 9. Clearly, almost all DEGs related to photosynthesis were down-regulated relative to mock inoculation at 12 hai. At 24 and 72 hai, down-regulation had ceased. However, down-regulation began again at 144 hai, reaching the maximum at 288 hai.
Figure 9.
Down-regulation of genes in photosynthesis-related pathways in response to P. pachyrhizi infection in the incompatible interaction. The bioinformatics program MapMan was used to visualize the effect of P. pachyrhizi infection on photosynthetic pathways over time. DEGs down-regulated relative to the mock-inoculated plants are green, while those expressed similarly to the mock-inoculated plants are white.
DISCUSSION
Microscopic Observations of P. pachyrhizi Infection
To better understand the interactions underlying the gene expression response in soybean leaves after P. pachyrhizi infection, we performed a time-course experiment from early infection through symptom development and sporulation in compatible and incompatible interactions mediated by the Rpp3 resistance gene. We monitored the growth and development of two P. pachyrhizi isolates (compatible interaction and incompatible interaction) microscopically and by qRT-PCR, and microarrays were used to assess soybean gene expression. In general, the developmental rate of appressoria, infection hyphae, and intercellular hyphae for these two isolates on the soybean cv Ankur were consistent with previous microscopic studies (Bonde et al., 1976; McLean, 1979; Koch et al., 1983). However, development of the avirulent isolate HW94-1 and the virulent isolate TW80-2 was slightly different at 12 hai. The lower number of penetrations and infection hyphae produced by the virulent TW80-2 showed that it did not carry out early infection processes as quickly as the avirulent HW94-1. However, TW80-2 did not continually lag behind HW94-1, because the numbers of appressoria, infection hyphae, and intercellular hyphae were similar for both isolates at 24 and 48 hai. The first detection of haustoria at 72 hai indicates that they were developing rapidly between 48 and 72 hai, which is somewhat later than the 24 to 48 hai that was reported previously (Deverall et al., 1977; McLean, 1979; Koch et al., 1983).
After haustoria were detected, the rate of fungal proliferation changed dramatically, and a clear distinction between the growth of TW80-2 and HW94-1 became evident. At 72 hai, virulent TW80-2 produced significantly more haustoria than avirulent HW94-1, and there were more necrotic and discolored cells producing autofluorescence in the incompatible interaction. The autofluorescence was not observed prior to the 72-hai time point and was likely due to the accumulation of phenolic compounds as part of a mounting defense response. These results indicate that Rpp3-mediated resistance was active at 72 hai and acting to limit the growth of HW94-1. Subsequently, HW94-1 elicited more extensive discolored, autofluorescent, and necrotic cells and produced clearly fewer intercellular hyphae than TW80-2 (compare the incompatible interaction in Fig. 3G with the compatible interaction in Fig. 3H), demonstrating that Rpp3 resistance was activated as HW94-1 attempted to grow.
In the compatible interaction, TW80-2 made a developmental transition from slow to rapid growth between 72 and 144 hai. The time that this transition occurred in the compatible interaction also correlated well with the time at which the defense response became more prominent in the incompatible interaction, demonstrating that the Rpp3 resistance response increased as HW94-1 also attempted to make this transition (i.e. more cells became engaged in the defense response). Differential P. pachyrhizi infection kinetics in compatible and incompatible interactions are consistent with previous reports (Marchetti et al., 1975; McLean, 1979; Keogh et al., 1980; Pua and Ilag, 1980), and they are similar to changes in infection kinetics between the incompatible and compatible interactions reported for poplar rust (Rinaldi et al., 2007) and wheat stripe rust (Coram et al., 2008).
Biphasic Gene Expression in Response to P. pachyrhizi Infection
The timeline for P. pachyrhizi growth and development discussed above provides a framework within which to view the complex gene expression changes that occurred during compatible and incompatible interactions in soybean. As we reported previously (van de Mortel et al., 2007), P. pachyrhizi isolates induce a striking biphasic response in soybean regardless of interaction type. We have now observed this response among interactions of three different soybean genotypes (Embrapa 48, PI230970, and Ankur [PI462312]) with three different P. pachyrhizi isolates (a Brazilian field isolate, HW94-1, and TW80-2). This biphasic response is characterized by strong differential gene expression at 12 hai, followed by the 24- to 48-hai period in which gene expression returns to the approximate level of the mock-inoculated plants. In our study, we could document that this first burst of differential gene expression corresponded to the early infection processes of appressorium formation and penetration of epidermal cells. Interestingly, the development of intercellular hyphae was coincident with gene expression profiles that returned to mock-inoculated levels. Differential gene expression appeared to be more extensive in the HW94-1-infected plants during this period (Fig. 4). However, as noted above, the formation of infection hyphae in TW80-2 was delayed relative to HW94-1 at 12 hai, while the two isolates appeared to develop with similar kinetics at later time points through 48 hai. Based on this observation, it is possible that the apparently stronger differential gene expression in response to HW94-1 at 12 hai was not due to Rpp3-mediated defenses but rather to a slight lag in the development of TW80-2 during the early infection processes. An alternative explanation for the stronger response to HW94-1 at 12 hai is that there could be some level of AvrRpp3 recognition in Rpp3 plants.
Following the early burst of gene expression is a quiescent period from 24 to 48 hai in which P. pachyrhizi continues early infection processes, but it does not elicit strong differential gene expression within soybean. During this time, the numbers of appressoria, infection hyphae, and intercellular hyphae plateaued for both HW94-1 and TW80-2. It is interesting that both P. pachyrhizi isolates cause such pronounced responses at 12 hai, yet gene expression returns nearly to normal by 24 hai. These observations suggest that P. pachyrhizi elicits a nonspecific defense response similar to PTI that was noneffective in stopping the pathogen. P. pachyrhizi is then able to suppress or evade early defense responses upon the development of intercellular hyphae and grow undetected after the initial penetration into the leaf mesophyll.
Our integrated microscopy-microarray approach identified the 72-hai time point as the benchmark for dynamic changes in P. pachyrhizi growth and development as well as the beginning of the second phase of soybean gene expression changes. The key factors influencing these changes were most likely related to the appearance of haustoria. At 72 hai, we found a second phase of differential gene expression developing in soybean leaves inoculated with HW94-1 but not the TW80-2 isolate. Unlike the first phase of differential gene expression, TW80-2 development was not delayed relative to HW94-1, because it had actually formed more haustoria than HW94-1. As the time course continued from this point, TW80-2 developed more rapidly and extensively in the Rpp3 plants than HW94-1. The stronger differential gene expression elicited by HW94-1 during this second phase is consistent with the activation of Rpp3-mediated resistance. In a previous study, we showed that P. pachyrhizi altered soybean gene expression in a biphasic manner and that the second phase of gene expression occurred earlier and at a higher magnitude in plants that carried the Rpp2 resistance gene (van de Mortel et al., 2007). That second phase of gene expression also began at 72 hai and occurred more rapidly and with a greater magnitude in the incompatible interaction. Given the similar kinetics of the Rpp3 and Rpp2 resistance responses, we infer that Rpp2-mediated resistance is also activated as the haustoria form.
The formation of haustoria by rust fungi results in increased surface area in close contact to the host cell plasma membrane. Rusts acquire nutrients from host cells through the haustoria and produce secreted proteins that are trafficked from haustoria into the plant cells (Staples, 2000, 2001; Voegele and Mendgen, 2003; Catanzariti et al., 2006). The set of secreted fungal proteins known as effectors that enter plant cells presumably carry out functions needed to promote nutrient acquisition and suppress host defense responses (Ellis et al., 2007b). Some fungal effectors also serve as avirulence determinants when they or their activities are recognized by corresponding resistance proteins in the plant (Stergiopoulos and de Wit, 2009). The induction of Rpp3-mediated resistance in Ankur coinciding with the formation of haustoria suggests that the putative AvrRpp3 product is produced by HW94-1, trafficked into the Ankur cells, and detected directly or indirectly by the Rpp3 resistance protein. AvrRpp3 has been neither genetically defined nor cloned, and similarly, the Rpp3 resistance gene has not been identified. Thus, it is currently not possible to determine if AvrRpp3 actually is expressed in haustoria, trafficked into host cells, and recognized intracellularly. Alternatively, the putative AvrRpp3 product could be secreted into the apoplast or extrahaustorial matrix and recognized by an Rpp3 protein that is localized to the surface of the cell (Stergiopoulos and de Wit, 2009). Interestingly, Rpp3 maps to a genetic interval containing several nucleotide-binding site Leu-rich repeat resistance gene analogs (Hyten et al., 2009). If Rpp3 is indeed a nucleotide-binding site Leu-rich repeat protein, then it would likely mediate the recognition of AvrRpp3 inside plant cells. We note that HW94-1 continues to grow and develop, albeit at a much reduced rate relative to TW80-2, demonstrating that Rpp3 resistance is not completely effective in immediately halting infection (Fig. 2A). The slow growth and development of HW94-1 in Ankur leads to continued and increasing differential gene expression through 288 hai. If Rpp3 resistance were 100% effective, one could expect that fungal growth would completely cease and that the resistance-associated gene expression response would be more transient.
Additional evidence that the earlier second phase in the incompatible interaction was specific to Rpp3 comes from the interactions that HW94-1 and TW80-2 have with other cultivars. HW94-1 is virulent on cultivars such as Williams, resulting in tan lesions, and TW80-2 is avirulent on Rpp4 genotypes such as PI459025B, resulting in RB lesions (Bonde et al., 2006). These observations suggest that the differential responses to HW94-1 and TW80-2 beginning at 72 hai were most likely specific to the Rpp3 resistance response. However, because HW94-1 and TW80-2 are not isogenic, we must also entertain the possibility that factors other than the AvrRpp3-Rpp3 interaction contributed to the differential responses observed on Ankur plants. Plant pathogenic fungi produce an array of effectors that can vary from one isolate to another, which could affect the interactions in isolate-specific ways.
A few studies now have investigated gene expression in soybean or the related species Glycine tomentella in response to P. pachyrhizi (Panthee et al., 2007, 2009; van de Mortel et al., 2007; Choi et al., 2008; Soria-Guerra et al., 2010a, 2010b; Tremblay et al., 2010). Of these, studies by van de Mortel et al. (2007) and Soria-Guerra et al. (2010a) used time courses that were extensive enough to potentially observe the biphasic response. The biphasic response in soybean is supported by our previous study, which was conducted with a different P. pachyrhizi isolate and with different soybean germplasm (van de Mortel et al., 2007). Gene expression in response to P. pachyrhizi was also monitored in G. tomentella over a time course spanning 12 to 72 hai (Soria-Guerra et al., 2010a). The authors observed similar numbers of DEGs at 12, 24, 48, and 72 hai, and there was extensive overlap in gene expression between compatible and incompatible interactions and between time points (i.e. a large drop in the number of differentially expressed genes was not observed at 24 hai compared with 12 hai, as we observed [van de Mortel et al., 2007; this study]). We performed hierarchical clustering and generated heat maps using the fold change data provided for the G. tomentella experiment and found that few G. tomentella genes were expressed in a biphasic manner (data not shown). These observations suggest that the first phase of the biphasic response to P. pachyrhizi may be much less pronounced or nonexistent in G. tomentella. The lack of a response specific to the incompatible interaction in G. tomentella may be due to the experimental time course. If the resistance response were activated beginning at 72 hai or later, as in our study, then major differences in gene expression would not have been found in the compatible and incompatible interactions in the studies reported by others. It would be interesting to perform a microscopic analysis of compatible and incompatible interactions to determine the kinetics of P. pachyrhizi growth and development in other Glycine species. This particular G. tomentella resistance response is not accompanied by macroscopic signs (Soria-Guerra et al., 2010a), so it is not possible to visually determine if it is elicited early or later as the haustoria form.
Nature of the Differentially Expressed Genes
We divided the soybean DEGs into two basic sets: (1) 1,799 DEGs expressed in response to HW94-1 and TW80-2 (common genes); and (2) 6,647 DEGs expressed in response to HW94-1 alone (unique genes). Only 27 DEGs were identified as unique in response to TW80-2. Therefore, the genes in Ankur that responded to TW80-2 were essentially a subset of those genes that responded to HW94-1. More interestingly, the very large set of unique genes provides a glimpse at a very complex resistance response mediated by Rpp3.
The common and unique sets of genes both exhibited biphasic expression patterns. The majority (75%) of common differentially expressed genes were up-regulated during the compatible and incompatible interactions, indicating that they had the most robust response to P. pachyrhizi (Fig. 6A). In contrast, only 41% of the DEGs unique to HW94-1 were up-regulated relative to the mock-inoculated plants (Fig. 6B). When the expression of annotated transcription factors was examined, approximately 55% were up-regulated among the common genes (Fig. 7A), whereas among the unique genes, only 33% were up-regulated (Fig. 7B). Therefore, there appears to be a clear difference in the expression profiles of genes that are differentially expressed in both interactions versus those unique to the incompatible interaction.
The GO categories related to photosynthesis were overrepresented among genes unique to the incompatible interaction (Supplemental Table S2). The expression profiles of these genes show that they are exclusively down-regulated in response to HW94-1 (Fig. 6B). The relationship between the photosynthetic machinery and plant defense is not well understood. There are many observations that the expression of genes related to photosynthesis is down-regulated rapidly in response to both virulent and avirulent pathogens (Bolton, 2009). Recently, Bilgin et al. (2010) compared plant microarray data from 22 sources of biotic stress in eight different plant species and found decreased expression of photosynthesis-related genes regardless of pathogen or plant species. These observations show that plant cells undergoing resistance responses or pathogen attack rapidly down-regulate the expression of photosynthesis-related genes. This may allow plants to better allocate resources to defend against pathogen attack (Bolton, 2009).
Our data for soybean rust are in agreement with those other plant-pathogen interactions. Furthermore, our data demonstrate how the down-regulation of photosynthetic genes is a feature of both nonspecific and specific defense reactions. At 12 hai, during the nonspecific phase of the interaction, there is a dramatic decrease in the expression of photosynthesis-related genes in response to HW94-1, and even though these genes did not meet the rigorous significance cutoff in the TW80-2 response, there was a general trend for their down-regulation in the compatible interaction. This initial response is akin to PTI responses, which have been associated with decreased expression of photosynthesis-related genes (Bolton, 2009). In both interactions, the expression of photosynthesis-related genes returned to mock-inoculation levels, but beginning at 144 hai, there was a second, sustained down-regulation of these genes in response to HW94-1. In the TW80-2-infected plants, the expression of these genes also trended downward, but at later time points and to a much smaller extent. The down-regulation of these genes correlated well with the appearance of RB lesions surrounded by intensely chlorotic regions by 12 d after inoculation in the incompatible interaction, while tan lesions that developed in the compatible interaction were surrounded by green or slightly chlorotic tissue (Fig. 1). The stronger repression of photosynthesis-related genes in the incompatible (HW94-1) interaction suggests that TW80-2 may modulate the response of the photosynthesis-related genes in the compatible interaction or simply does not elicit massive changes in their expression. Active modulation of photosynthesis-related genes would be consistent with the green island effect characterized by sustained photosynthesis in tissues infected with biotrophic fungi (Schulze-Lefert and Vogel, 2000).
Tremblay et al. (2010) observed significant down-regulation of photosynthesis-related genes in the compatible response in soybean to P. pachyrhizi. The tissues, examined from later stages of infection development, were largely discolored and possibly undergoing cell death and senescence responses. Long-term decreases in the expression of photosynthesis-related genes in compatible interactions would be consistent with observations of actual photosynthesis in P. pachyrhizi-infected soybean leaves. As P. pachyrhizi infection progresses, reduced photosynthesis and assimilation of CO2 occur, due in part to defoliation and reduction of the green leaf area index as lesions form (Kumudini et al., 2008a). In addition, P. pachyrhizi also reduces the efficiency of photosynthesis in the remaining green tissues that visually appear to be healthy (Kumudini et al., 2008b). This suggests that the down-regulation of photosynthesis observed by Tremblay et al. (2010) is a consequence of disease and not a resistance response. Looking at earlier time points in the compatible interaction, we did not find significant down-regulation of photosynthesis-related genes, most likely because the number of cells undergoing cell death was a relatively small proportion of the cells examined and their gene expression changes were diluted. In contrast, the down-regulation of photosynthesis-related genes in the incompatible interaction was due directly to the resistance response, with a much larger number of cells undergoing the hypersensitive response and cell death. It will be interesting to further investigate the relationship between P. pachyrhizi-soybean interactions and photosynthetic processes.
Another interesting class of genes is the transcription factors. Comparison across transcription factor classes on the common and unique-to-HW94-1 gene lists reveals very distinct profiles of expression. A few examples of this are the WRKY and NAC transcription factors that are nearly all up-regulated on both lists. The AUX-IAA-ARF and MYB-HD classes appear to be exclusively down-regulated on both gene lists. The AP2-EREBP transcription factors are mostly up-regulated on the common gene list, but on the unique-to-incompatible list, about half of them are up-regulated while the other half are down-regulated. The differing patterns of expression suggest coregulation among the different transcription factor families. The AUX-IAA-ARF and MYB-HD transcription factor classes are mostly involved in regulating plant growth and development. The down-regulation of these transcription factors is consistent with the idea that growth and development are negatively affected during pathogen defense (Bolton, 2009). The increased expression of WRKY and NAC transcription factors implies that their increased expression is needed to control defense responses either through positive or negative regulatory functions. This is consistent with the prominent role of these transcription factors in regulating plant defense responses (Van Verk et al., 2009). The AP2-EREBP transcription factors may be involved in plant defense or plant growth and development. The extensive reprogramming of host gene expression is consistent with the large number of transcription factors that are differentially regulated. The implication is that resistance to P. pachyrhizi depends on a complex and interwoven network of genes. These data combined with transcriptome data from loss-of-function experiments for R genes and downstream signaling genes will provide key resources needed for network modeling of soybean defense responses.
CONCLUSION
Analysis of soybean defense networks against P. pachyrhizi and other pathogens requires data sets that effectively consider the growth and development of the invading pathogen relative to the host responses. Our results show that microscopic analysis of P. pachyrhizi growth and development coupled with gene expression measurements provide insight into the responses observed in soybean at various points during the infection process. The results also show that P. pachyrhizi isolates can develop differently and that differences in the timing of development can influence the apparent response of the host plant. In addition, we saw that the development of haustoria represents a milestone in P. pachyrhizi-soybean interactions that was followed by rapid proliferation of the fungus in the compatible interaction. In the incompatible interaction, formation of haustoria marked the point at which development of the avirulent P. pachyrhizi isolate HW94-1 was clearly impaired. A plausible explanation for these observations is that an effector produced by HW94-1 haustoria is secreted and recognized directly or indirectly by Rpp3, resulting in defense responses that limit infection. P. pachyrhizi growth and development as well as the concomitant soybean responses all are dynamic processes. Information on the kinetic relationships of these processes is helping to further our understanding of the gene networks in soybean and P. pachyrhizi that interface to mediate the outcomes of the interaction.
MATERIALS AND METHODS
Experimental Design and Inoculation
All inoculations were performed in the U.S. Department of Agriculture-Agricultural Research Service Foreign Disease-Weed Science Research Unit Biological Safety Level-3 Plant Pathogen Containment Facility at Fort Detrick, Maryland (Melching et al., 1983). The TW80-2 isolate was originally collected in 1980 from infected soybean (Glycine max) leaves in a field plot at the Asian Vegetable Research and Development Center, in Taipei, Taiwan, and the HW94-1 isolate was collected in 1994 from infected soybean leaves in Oahu, Hawaii (Bromfield, 1984; Hartwig, 1986; Bonde et al., 2006; Hyten et al., 2009). Urediniospores of the two Phakopsora pachyrhizi isolates TW80-2 and HW94-1 were removed from liquid nitrogen, heat shocked at 40°C for 5 min, and hydrated at 100% relative humidity at room temperature for 16 h. Spores were suspended in sterile distilled water containing 0.01% (v/v) Tween 20, filtered through a 53-μm pore-size screen, and adjusted to a concentration of 5 × 105 spores mL−1 by means of a hemacytometer. The Rpp3-containing soybean cv Ankur (PI462312) used in this experiment develops resistant RB lesions when infected with HW94-1 and susceptible tan lesions when infected with TW80-2. Two plants per pot were inoculated at the V3 growth stage (Fehr and Caviness, 1977) by spraying approximately 10 mL of urediniospore solution with an atomizer at 20 p.s.i. onto the adaxial surface of the leaves. The same solution minus urediniospores was used for the mock inoculations. Following the P. pachyrhizi or mock inoculation, the plants were placed in dew chambers for 12 h at 20°C, after which they were moved to the greenhouse at 20°C. Supplemental illumination was provided by 1,000-W Metalarc lights (Sylvania) spaced 0.6 m apart above the bench. The third trifoliolate leaves of two plants were collected for microarray analysis for the first 12 d of infection. The experiment followed a randomized complete block design with the three replicates as blocks and with a full factorial treatment structure with two treatment factors: time (six levels) and infection type (HW94-1, TW80-2, or mock inoculation).
Tissue Collection and RNA Isolation
The three leaflets of the third trifoliolate leaves of two plants (six leaflets total) were collected at 12, 24, 72, 144, 216, and 288 hai. One leaf was preserved for microscopic observations (see below), and the remaining leaves were immediately frozen in liquid nitrogen and stored at −80°C. Leaf tissue was ground in liquid nitrogen, and RNA was extracted from approximately 200 mg using 1 mL of Tri Reagent (Molecular Research Center). RNA samples were further purified by an overnight precipitation at −20°C in 2 m (final concentration) lithium chloride (Ausubel et al., 1994) followed by RNeasy column purification (Qiagen) and elution in 30 μL of diethyl pyrocarbonate-treated water.
Microscopic Observations of ASR Infection Structures
At each sampling time (12, 24, 48, 72, 96, 120, 144, 216, and 288 hai), three leaflets were collected for each infection type (HW94-1, TW80-2, and mock inoculated) and fixed in Farmer’s solution (3:1 [v/v] 100% ethanol:glacial acetic acid). Samples were prepared by harvesting 0.5-cm2 tissue samples from one leaflet of the third trifoliate during the first 9 d of infection, stained for 48 h in a clearing/staining solution containing chlorazol black (Keane et al., 1988), and cleared in a saturated solution of chloral hydrate for several days. In order to obtain microscopic data for the visualization of autofluorescence, tissues were fixed in boiling 95% ethanol and cleared in saturated chloral hydrate (Heath, 1984). Specimens were mounted in 50% glycerol and examined via differential interference contrast microscopy. Specimens prepared for autofluorescence were examined using an epifluorescence microscope (Leitz Fluovert; exciter filter BP 420–490, dichroic mirror RKP 510, barrier filter LP 520).
Data were taken on individual fungal colonies developing in the infected leaflets. Clumps of urediniospores or colonies growing close enough together that their mycelia were in close contact were not included in the analysis. Colonies could be examined from the appressoria that developed on the adaxial leaflet surface down through the palisade mesophyll to the stellate layer in the middle of the leaflet. P. pachyrhizi colonies were restricted to these tissues during the first 96 hai, and all quantitative data were obtained while viewing from the adaxial surface. Additional observations of fungal development within the spongy mesophyll were obtained by viewing leaf tissues from the abaxial surface.
Assessment of Fungal mRNA Accumulation
Fungal growth was assessed by quantifying the constitutively expressed P. pachyrhizi α-tubulin gene by TaqMan qRT-PCR as described previously (van de Mortel et al., 2007). The iScript One-Step RT-PCR kit for probes (Bio-Rad) was used according to the manufacturer’s protocol with 50 ng of RNA, 300 nm final concentration primers (forward, 5′-CCAAGGCTTCTTCGTGTTTCA-3′; reverse, 5′-CAAGAGAAGAGCGCCAAACC-3′), and 150 nm probe (5′-FAM-TCGTTTGGAGGCGGACTGGTTCA-3′ Black Hole Quencher BHQ-1 (Integrated DNA Technologies) in the following RT-PCR program: cDNA synthesis for 10 min at 50°C, iScript reverse transcription inactivation for 5 min at 95°C, PCR cycling at 95°C for 10 s, and data collection for 30 s at the extension temperature of 60°C for 45 cycles. Expression data were normalized to the soybean ubiquitin-3 gene (accession no. gi 456713, dbj D28123.1; forward primer, 5′-GTGTAATGTTGGATGTGTTCCC-3′; reverse primer, 5′-ACACAATTGAGTTCAACACAAACCG-3′; extension temperature = 65°C and 35 amplification cycles; Trevaskis et al., 2002), which showed no evidence for differential expression in our experiments.
Microarray Labeling, Hybridization, and Scanning
RNA samples were adjusted to a concentration of 0.6 μg μL−1, and soybean mRNA transcript abundance was measured using GeneChip Soybean Genome Arrays (Affymetrix). RNA concentration and quality were determined using a NanoDrop spectrophotometer (NanoDrop Technologies) and by RNA Nano LabChip on a 2100 Bioanalyzer (Agilent Technologies). All steps in labeling, hybridization, and scanning were performed at the Iowa State University GeneChip Facility. The synthesis of labeled target copy RNA (cRNA) used 5 μg of total RNA and was performed using the GeneChip One-Cycle Target Labeling and Control Reagents kit (Affymetrix) according to the manufacturer’s instructions. Ten micrograms of fragmented cRNA was hybridized to GeneChip Soybean Genome Arrays (Affymetrix) according to the manufacturer’s instructions. The quality of fragmented cRNA was verified on an Agilent 2100 Bioanalyzer with an RNA Nano LabChip. Washes were performed using the EukGE-WS2v5_450 washing protocol, and microarrays were scanned with a GCS3000 7G scanner (Affymetrix).
Statistical Analysis of Microarray Data
The base 2 logarithm of MAS5.0 signals was median centered so that the median log-scale expression measure for each GeneChip was zero. Linear model analysis of these normalized log-scale expression measures was performed separately for each gene using version 9.2 of SAS (SAS Institute). Each linear model included fixed replication effects and fixed effects for times, infection types, and interactions between times and infection types. As part of each linear model analysis, F tests were conducted for infection type main effects, infection type-by-time interactions, and all possible comparisons of infection types at each time point. These tests were used to determine if there was significant evidence of an expression difference between infection types when averaging over time, whether there was significant evidence that the pattern of expression over time differed with infection type, and whether infection types differed significantly within individual time points. Together, these tests were used to search for genes whose expression differed in some manner (either in level, pattern over time, or both) between infection types within each genotype. In addition, F tests based on contrasts of means were used to identify genes whose expression pattern over the entire time course differed between mock- and HW94-1 treated plants and/or between mock- and TW80-2 treated plants. A q value was computed for each F test P value using the method described by Storey and Tibshirani (2003). The q values were used to produce lists of differentially expressed genes with estimated FDRs of 0.01%. Hierarchical clustering in R programming language for statistical computing using the Agnes function was performed on the standardized base 2 logarithm of fold change in gene expression on various data sets according to test results. Hierarchical clustering using Pearson correlation with average linkage in cluster 3.0 was performed for clustering the base 2 logarithm of the fold change in gene expression of the P. pachyrhizi-regulated probe sets, and heat maps were displayed using Java TreeView (de Hoon et al., 2004; Saldanha, 2004).
Gene Annotation
DEGs were annotated using the SoyBase Affymetrix GeneChip Soybean Genome Array Annotation page (version 2) as described earlier (van de Mortel et al., 2007; http://www.soybase.org/AffyChip/). Briefly, target sequences from which probes were designed were compared with the predicted cDNAs from the soybean whole genome assembly (version 1.0; Schmutz et al., 2010) using BLASTN (Altschul et al., 1997). Soybean cDNA matches had a required minimum length of 100 bp, E value cutoff of 10−30, and 95% nucleotide identity. If no matching soybean cDNAs could be identified or multiple cDNAs from multiple genes could not be distinguished, the Affymetrix consensus sequence was used in place of the soybean cDNA for further analyses. The soybean predicted cDNAs, and when necessary the Affymetrix consensus sequences, were compared with the UniProt protein database (Apweiler et al., 2004) and predicted cDNAs from the Arabidopsis (Arabidopsis thaliana) genome (version 8; The Arabidopsis Information Resource [www.arabidopsis.org]) using BLASTX (E < 10−6; Altschul et al., 1997) and The Arabidopsis Information Resource GO and GO slim annotations (Berardini et al., 2004) for each Arabidopsis sequence identified. In order to cross-reference probes with soybean transcription factor families, custom Perl scripts were used to assign transcription factor categories from SoyDB (Wang et al., 2010) to corresponding probes based on the soybean cDNA sequence assigned to the probe. Fisher’s exact test with a Bonferroni correction was used to identify overrepresented GO categories or transcription factor classes (van de Mortel et al., 2007).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Probe sets corresponding to genes that were differentially expressed in response to P. pachyrhizi isolates HW94-1 or TW80-2 in Rpp3 plants.
Supplemental Table S2. Grouping of overrepresented GO biological function terms found in probe sets differentially expressed in response to P. pachyrhizi infection.
Acknowledgments
We thank Dr. Jiqing Peng for processing the RNA samples for GeneChip analysis, JoAnn Bowers for assistance with ASR inoculations, and Jaime Dittman for her contributions to this research. We also thank Dr. Mandy Kendrick for critical review of the manuscript.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. (2004) UniProt: the Universal Protein knowledgebase. Nucleic Acids Res 32: D115–D119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. (1994) Current Protocols in Molecular Biology. John Wiley & Sons, Hoboken, NJ [Google Scholar]
- Bari R, Jones JDG. (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Mundodi S, Reiser L, Huala E, Garcia-Hernandez M, Zhang P, Mueller LA, Yoon J, Doyle A, Lander G, et al. (2004) Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol 135: 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, DeLucia EH. (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33: 1597–1613 [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bolton MD. (2009) Primary metabolism and plant defense: fuel for the fire. Mol Plant Microbe Interact 22: 487–497 [DOI] [PubMed] [Google Scholar]
- Bonde MR, Melching JS, Bromfield KR. (1976) Histology of suscept-pathogen relationship between Glycine max and Phakopsora pachyrhizi, cause of soybean rust. Phytopathology 66: 1290–1294 [Google Scholar]
- Bonde MR, Nester SE, Austin CN, Stone CL, Frederick RD, Hartman GL, Miles MR. (2006) Evaluation of virulence of Phakopsora pachyrhizi and P. meibomiae isolates. Plant Dis 90: 708–716 [DOI] [PubMed] [Google Scholar]
- Bonferroni CE. (1935) Il calcolo delle assicurazioni su gruppi di teste. Studi in Onore del Professore Salvatore Ortu Carboni; Rome, pp 13–60 [Google Scholar]
- Bromfield KR. (1984) Soybean Rust. Monograph No. 11. American Phytopathological Society, St. Paul, MN [Google Scholar]
- Bromfield KR, Hartwig EE. (1980) Resistance to soybean rust and mode of inheritance. Crop Sci 20: 254–255 [Google Scholar]
- Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. (2006) Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YW, Chan KL. (1968) The breeding of ‘Tainung 3’ soybean. J Taiwan Agric Res 17: 30–35 [Google Scholar]
- Choi JJ, Alkharouf NW, Schneider KT, Matthews BF, Frederick RD. (2008) Expression patterns in soybean resistant to Phakopsora pachyrhizi reveal the importance of peroxidases and lipoxygenases. Funct Integr Genomics 8: 341–359 [DOI] [PubMed] [Google Scholar]
- Coram TE, Settles ML, Chen X. (2008) Transcriptome analysis of high-temperature adult-plant resistance conditioned by Yr39 during the wheat-Puccinia striiformis f. sp. tritici interaction. Mol Plant Pathol 9: 479–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. (2010) The plant hormones: their nature, occurrence, and functions. Davies PJ, , Plant Hormones. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 1–15 [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. (2004) Open source clustering software. Bioinformatics 20: 1453–1454 [DOI] [PubMed] [Google Scholar]
- Deverall BJ, Keogh RC, McLeod S. (1977) Responses of soybean to infection by, and to germination fluids from urediniospores of Phakopsora pachyrhizi. Trans Br Mycol Soc 69: 411–415 [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drǎghici S, Khatri P, Martins RP, Ostermeier GC, Krawetz SA. (2003) Global functional profiling of gene expression. Genomics 81: 98–104 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Dodds PN, Lawrence GJ. (2007a) Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol 45: 289–306 [DOI] [PubMed] [Google Scholar]
- Ellis JG, Dodds PN, Lawrence GJ. (2007b) The role of secreted proteins in diseases of plants caused by rust, powdery mildew and smut fungi. Curr Opin Microbiol 10: 326–331 [DOI] [PubMed] [Google Scholar]
- Fehr WR, Caviness CE. (1977) Stages of Soybean Development. Special Report 80. Iowa State University of Science and Technology, Iowa Agriculture and Home Economics Experiment Station, Ames, IA [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T. (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fisher RA. (1966) The Design of Experiments, Ed 8. Oliver and Boyd, Edinburgh [Google Scholar]
- Garcia A, Calvo ES, de Souza Kiihl RA, Harada A, Hiromoto DM, Vieira LG. (2008) Molecular mapping of soybean rust (Phakopsora pachyrhizi) resistance genes: discovery of a novel locus and alleles. Theor Appl Genet 117: 545–553 [DOI] [PubMed] [Google Scholar]
- Goellner K, Loehrer M, Langenbach C, Conrath U, Koch E, Schaffrath U. (2010) Phakopsora pachyrhizi, the causal agent of Asian soybean rust. Mol Plant Pathol 11: 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T. (1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- Hartwig EE. (1986) Identification of a fourth major gene conferring resistance to soybean rust. Crop Sci 26: 1135–1136 [Google Scholar]
- Hartwig EE, Bromfield KR. (1983) Relationships among three genes conferring specific resistance to rust in soybeans. Crop Sci 23: 237–239 [Google Scholar]
- He K, Guo AY, Gao G, Zhu QH, Liu XC, Zhang H, Chen X, Gu X, Luo J. (2010) Computational identification of plant transcription factors and the construction of the PlantTFDB database. Methods Mol Biol 674: 351–368 [DOI] [PubMed] [Google Scholar]
- Heath MC. (1984) Relationship between heat-induced fungal death and plant necrosis in compatible and incompatible interactions involving the bean and cowpea rust fungi. Phytopathology 74: 1370–1376 [Google Scholar]
- Hidayat OO, Somaatmadja S. (1977) Screening of soybean breeding lines for resistance to soybean rust (Phakopsora pachyrhizi Sydow). Soybean Rust Newsl. 1: 9–22 [Google Scholar]
- Hoefle C, Loehrer M, Schaffrath U, Frank M, Schultheiss H, Hückelhoven R. (2009) Transgenic suppression of cell death limits penetration success of the soybean rust fungus Phakopsora pachyrhizi into epidermal cells of barley. Phytopathology 99: 220–226 [DOI] [PubMed] [Google Scholar]
- Hyten DL, Smith JR, Frederick RD, Tucker ML, Song Q, Cregan PB. (2009) Bulked segregant analysis using the GoldenGate assay to locate the Rpp3 locus that confers resistance to soybean rust in soybean. Crop Sci 49: 265–271 [Google Scholar]
- Jones JDG, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Keane PJ, Limongiello N, Warren MA. (1988) A modified method for clearing and staining leaf-infecting fungi in whole leaves. Australas Plant Pathol 17: 37–38 [Google Scholar]
- Keogh RC, Deverall BJ, McLeod S. (1980) Comparison of histological and physiological-responses to Phakopsora pachyrhizi in resistant and susceptible soybean. Trans Br Mycol Soc 74: 329–333 [Google Scholar]
- Koch E, Ebrahim-Nesbat F, Hoppe HH. (1983) Light and electron-microscopic studies on the development of soybean rust (Phakopsora pachyrhizi Syd) in susceptible soybean leaves. Phytopathol Z 106: 302–320 [Google Scholar]
- Kumudini S, Godoy CV, Board JE, Omielan J, Tollenaar M. (2008a) Mechanisms involved in soybean rust-induced yield reduction. Crop Sci 48: 2334–2342 [Google Scholar]
- Kumudini S, Prior E, Omielan J, Tollenaar M. (2008b) Impact of Phakopsora pachyrhizi infection on soybean leaf photosynthesis and radiation absorption. Crop Sci 48: 2343–2350 [Google Scholar]
- La Camera S, Gouzerh G, Dhondt S, Hoffmann L, Fritig B, Legrand M, Heitz T. (2004) Metabolic reprogramming in plant innate immunity: the contributions of phenylpropanoid and oxylipin pathways. Immunol Rev 198: 267–284 [DOI] [PubMed] [Google Scholar]
- Loehrer M, Langenbach C, Goellner K, Conrath U, Schaffrath U. (2008) Characterization of nonhost resistance of Arabidopsis to the Asian soybean rust. Mol Plant Microbe Interact 21: 1421–1430 [DOI] [PubMed] [Google Scholar]
- Mackey D, McFall AJ. (2006) MAMPs and MIMPs: proposed classifications for inducers of innate immunity. Mol Microbiol 61: 1365–1371 [DOI] [PubMed] [Google Scholar]
- Marchetti MA, Uecker FA, Bromfield KR. (1975) Uredial development of Phakopsora pachyrhizi in soybeans. Phytopathology 65: 822–823 [Google Scholar]
- McLean RJ. (1979) Histological studies of resistance to soybean rust, Phakopsora pachyrhizi Syd. Aust J Agric Res 30: 77–84 [Google Scholar]
- McLean RJ, Byth DE. (1980) Inheritance of resistance to rust (Phakopsora pachyrhizi) in soybeans. Aust J Agric Res 31: 951–956 [Google Scholar]
- Meindl T, Boller T, Felix G. (2000) The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melching JS, Bromfield KR, Kingsolver CH. (1983) The plant pathogen containment facility at Frederick, Maryland. Plant Dis 67: 717–722 [Google Scholar]
- Monteros MJ, Missaoui AM, Phillips DV, Walker DR, Boerma HR. (2007) Mapping and confirmation of the ‘Hyuuga’ red-brown lesion resistance gene for Asian soybean rust. Crop Sci 47: 829–834 [Google Scholar]
- Nettleton D. (2006) A discussion of statistical methods for design and analysis of microarray experiments for plant scientists. Plant Cell 18: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle HJ, Byth DE, McLean R. (1979) Effect of rust (Phakopsora pachyrhizi) on soybean yield and quality in south-eastern Queensland. Aust J Agric Res 30: 883–893 [Google Scholar]
- Panthee DR, Marois JJ, Wright DL, Narváez D, Yuan JS, Stewart CN., Jr (2009) Differential expression of genes in soybean in response to the causal agent of Asian soybean rust (Phakopsora pachyrhizi Sydow) is soybean growth stage-specific. Theor Appl Genet 118: 359–370 [DOI] [PubMed] [Google Scholar]
- Panthee DR, Yuan JS, Wright DL, Marois JJ, Mailhot D, Stewart CN., Jr (2007) Gene expression analysis in soybean in response to the causal agent of Asian soybean rust (Phakopsora pachyrhizi Sydow) in an early growth stage. Funct Integr Genomics 7: 291–301 [DOI] [PubMed] [Google Scholar]
- Patil VS, Wuike RV, Thakare CS, Chirame BB. (1997) Viability of uredospores of Phakopsora pachyrhizi Syd. at different storage conditions. J Maharashtra Agric Univ 22: 260–261 [Google Scholar]
- Pua AR, Ilag LL. (1980) Ingress and pathogenic development of Phakopsora pachyrhizi Syd. in soybean. Philipp Agric 63: 9–14 [Google Scholar]
- Rafiqi M, Gan PH, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, Hardham AR, Dodds PN. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22: 2017–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravensdale M, Nemri A, Thrall PH, Ellis JG, Dodds PN. (2011) Co-evolutionary interactions between host resistance and pathogen effector genes in flax rust disease. Mol Plant Pathol 12: 93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi C, Kohler A, Frey P, Duchaussoy F, Ningre N, Couloux A, Wincker P, Le Thiec D, Fluch S, Martin F, et al. (2007) Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol 144: 347–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ. (2004) Java TreeView: extensible visualization of microarray data. Bioinformatics 20: 3246–3248 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert P, Vogel J. (2000) Closing the ranks to attack by powdery mildew. Trends Plant Sci 5: 343–348 [DOI] [PubMed] [Google Scholar]
- Singh BB, Thapliyal PN. (1977) Breeding for resistance to soybean rust in India. Ford RE, Sinclair JB, , Rust of Soybean: The Problem and Research Needs. INTSOY Series No. 12. College of Agriculture, University of Illinois at Urbana-Champaign, Urbana, IL, pp 62–65 [Google Scholar]
- Soria-Guerra RE, Rosales-Mendoza S, Chang S, Haudenshield JS, Padmanaban A, Rodriguez-Zas S, Hartman GL, Ghabrial SA, Korban SS. (2010a) Transcriptome analysis of resistant and susceptible genotypes of Glycine tomentella during Phakopsora pachyrhizi infection reveals novel rust resistance genes. Theor Appl Genet 120: 1315–1333 [DOI] [PubMed] [Google Scholar]
- Soria-Guerra RE, Rosales-Mendoza S, Chang S, Haudenshield JS, Zheng D, Rao SS, Hartman GL, Ghabrial SA, Korban SS. (2010b) Identifying differentially expressed genes in leaves of Glycine tomentella in the presence of the fungal pathogen Phakopsora pachyrhizi. Planta 232: 1181–1189 [DOI] [PubMed] [Google Scholar]
- Staples RC. (2000) Research on the rust fungi during the twentieth century. Annu Rev Phytopathol 38: 49–69 [DOI] [PubMed] [Google Scholar]
- Staples RC. (2001) Nutrients for a rust fungus: the role of haustoria. Trends Plant Sci 6: 496–498 [DOI] [PubMed] [Google Scholar]
- Stergiopoulos I, de Wit PJ. (2009) Fungal effector proteins. Annu Rev Phytopathol 47: 233–263 [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay A, Hosseini P, Alkharouf N, Li S, Matthews BF. (2010) Transcriptome analysis of a compatible response by Glycine tomentella to Phakopsora pachyrhizi infection. Plant Sci 179: 183–193 [Google Scholar]
- Trevaskis B, Wandrey M, Colebatch G, Udvardi MK. (2002) The soybean GmN6L gene encodes a late nodulin expressed in the infected zone of nitrogen-fixing nodules. Mol Plant Microbe Interact 15: 630–636 [DOI] [PubMed] [Google Scholar]
- van de Mortel M, Recknor JC, Graham MA, Nettleton D, Dittman JD, Nelson RT, Godoy CV, Abdelnoor RV, Almeida ÁMR, Baum TJ, et al. (2007) Distinct biphasic mRNA changes in response to Asian soybean rust infection. Mol Plant Microbe Interact 20: 887–899 [DOI] [PubMed] [Google Scholar]
- Van Verk MC, Gatz C, Linthorst HJM. (2009) Transcriptional regulation of plant defense responses. Adv Bot Res 51: 397–437 [Google Scholar]
- Voegele RT, Mendgen K. (2003) Rust haustoria: nutrient uptake and beyond. New Phytol 159: 93–100 [DOI] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. (2008a) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Stacey G. (2008b) Chitin signaling and plant disease resistance. Plant Signal Behav 3: 831–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Libault M, Joshi T, Valliyodan B, Nguyen HT, Xu D, Stacey G, Cheng J. (2010) SoyDB: a knowledge database of soybean transcription factors. BMC Plant Biol 10: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise RP, Caldo RA, Hong L, Shen L, Cannon EK, Dickerson JA. (2007) BarleyBase/PLEXdb: a unified expression profiling database for plants and plant pathogens. Methods Mol Biol 406: 347–363 [DOI] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]