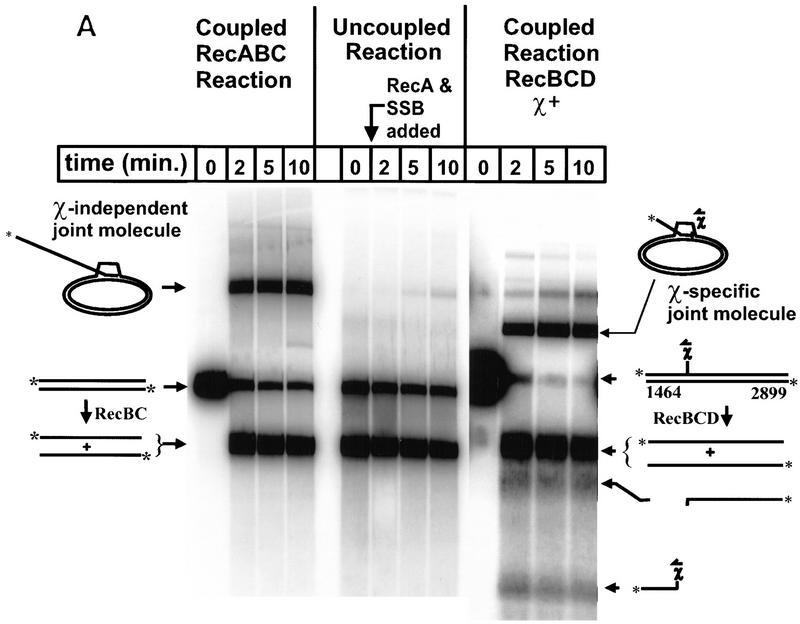
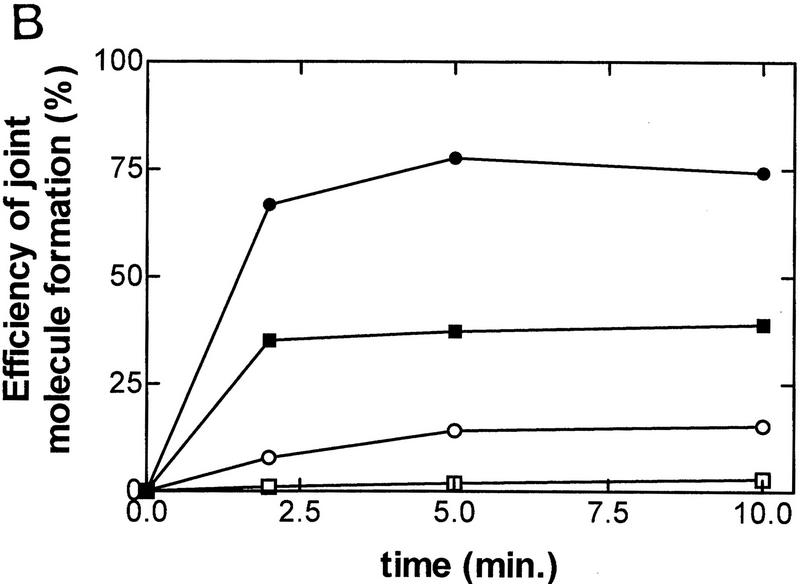
Figure 2.
Efficient formation of joint molecules in RecABC reactions requires the concerted action of RecA protein and RecBC enzyme. Linearized dsDNA was labeled at the 5′ end with 32P. The positions of labeled dsDNA substrate, ssDNA unwinding products, and joint molecules are indicated. Reaction conditions are described in Materials and Methods. (A) The coupled reactions were initiated by addition of either RecBC enzyme or RecBCD enzyme, in the presence of both RecA protein and SSB. In the uncoupled reaction, RecBC and RecBCD enzymes were omitted, heat-denatured DNA was substituted for dsDNA, and the reaction was initiated by the addition of a mixture of RecA and SSB proteins. The DNA substrates were XbaI-restricted χ0 pSKPB10, and HindIII-restricted χ+F (for the reaction with RecBCD). The sizes (in nucleotides) of fragments generated by RecBCD enzyme are indicated. (B) The efficiency of joint molecule formation is compared graphically for coupled vs. uncoupled reactions. (█) RecBC, coupled; (□) uncoupled (denatured DNA); (●) RecBCD, coupled χ-specific; (○) RecBCD, coupled full length. The efficiency of joint molecule formation is defined as the percentage of the specific ssDNA generated that appears in the respective joint molecule. The data for the coupled RecBC enzyme and uncoupled reactions represent an average of duplicates; there was no significant variation between replicates. Joint molecule formation from χ-specific fragments and full-length ssDNA produced by RecBCD enzyme is also shown for comparison.