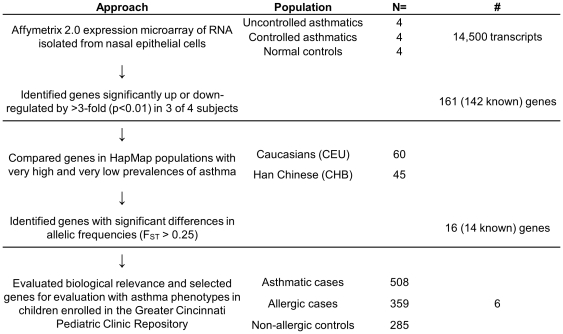
Figure 1. Novel unbiased approach to identify candidate asthma susceptibility genes.
The approach consisted of three stages with an evaluation of RNA expression of 14,500 genes in nasal epithelial samples in stage 1. Next, 142 known genes with >3-fold difference between uncontrolled asthmatics and non-allergic controls (P<0.05) were taken forward to stage 2. In stage 2, using HapMap data, the allelic frequencies of Caucasians (Utah residents of European ancestry; CEU) and the Han Chinese (Beijing, China; CHB) were compared and 14 known genes with a fixation index (FST)>0.25 were identified. Six of these genes, which mapped to chromosomal regions that had been linked to asthma previously, were included in the next phase. In stage 3, tagging SNPs including all CEU and YRI (Yoruban residents of Ibadan, Nigeria) SNPs with minor allele frequency less than 0.05 in the 6 genes were genotyped in children with asthma, allergic rhinitis or atopic dermatitis without asthma, and in non-allergic control children using a custom Illumina Golden Gate SNP Chip. Seven SNPs in a single gene, KIF3A, were significantly associated with asthma after adjusting for multiple comparisons (p-value<0.0006).