Figure 1.
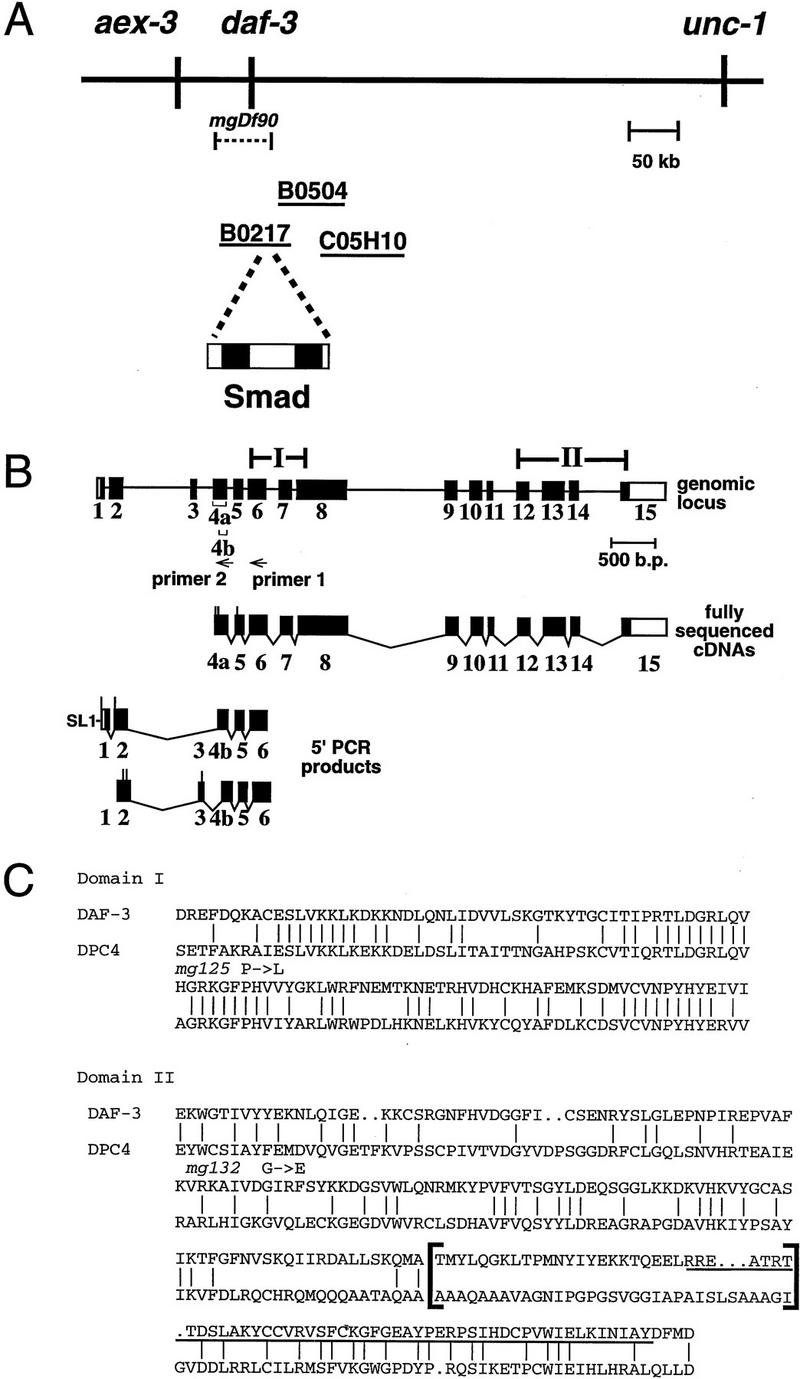
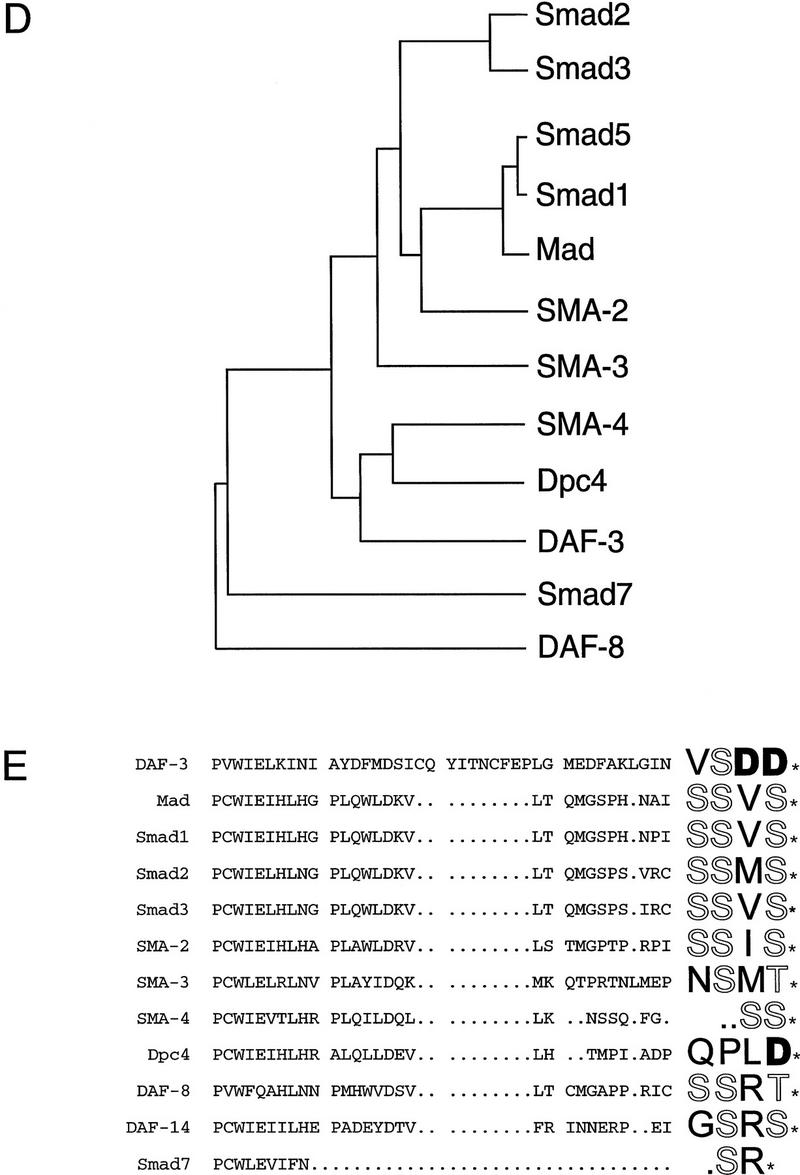
daf-3 encodes a Smad protein most closely related to DPC4. (A) daf-3 cloning. daf-3 was genetically mapped to a region on the X chromosome between aex-3 and unc-1 (A. Koweek and G. Patterson, unpubl.). B0217 complements daf-3 mutants in transformation rescue experiments; B0504 and C05H10 do not. mgDf90 is a deletion that removes all of daf-3. (B) Structure of daf-3-coding region. At the top is the exon/intron structure of daf-3; coding exons are solid boxes, noncoding regions are open boxes, and lines are introns. Conserved Smad domains I and II are indicated. The three classes of cDNA are shown, and 5′ ends are indicated by vertical lines. The accession numbers for the cDNAs shown are, top to bottom: AF005205, AF005206, and AF005207. (C) Protein sequence alignment of C. elegans DAF-3 and its closest homolog, human DPC4, in the Smad conserved domains I and II. Dots indicate gaps introduced to maximize alignment. The Smad mutational hot spot is underscored, and the insertion in DAF-3 and DPC4 relative to other Smads is bracketed. In addition to mg125 and mg132, seven other daf-3 alleles were sequenced in the hot spot; none of them contains a mutation. Alleles sequenced were mg91, mg93, mg105, mg121, mg126, mg133 (isolated by A. Koweek and G. Patterson, unpubl.) and sa205 (Thomas et al. 1993). (D) Relationship of DAF-3 domain I to other Smads. Lineup was performed with the program pileup (Genetics Computer Group 1994) using amino acids 137–245 of DAF-3 (GenBank accession no. AF005205) and corresponding residues of the other Smads. (E) Comparison of carboxyl termini of Smads. The final 28–44 residues are shown. Residues that are phosphorylated by receptor (in Smad1 and Smad2) or similar residues in similar positions (in other Smads) are shown in outline. Aspartates in similar positions are shown in boldface type.
