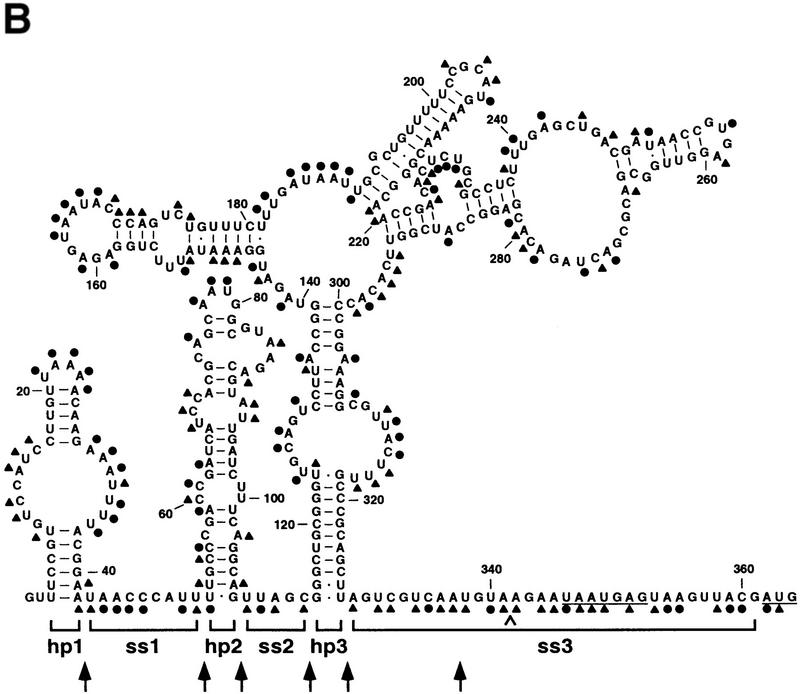
Figure 3.

Alkylation analysis of the E. coli rne 5′ UTR. (A) Representative analysis of a segment of the E. coli rne 5′ UTR by chemical alkylation. Total cellular RNA was isolated from an exponential culture of E. coli strain CH1828 containing plasmid pEZ101 after treating an aliquot of the culture with DMS. In addition, samples of RNA extracted from an untreated culture were alkylated in vitro with DMS or CMCT. Sites of alkylation within the ez1 5′ UTR were mapped by primer extension using AMV reverse transcriptase and various 5′ end-labeled DNA primers. The resulting extension products were then analyzed by gel electrophoresis beside sequencing ladders that were generated by extension of the same 5′ end-labeled primer on an ez1 DNA template. The sequencinglanes (lanes G,A,U,C) are labeled to indicatethe sequence of the RNA, not the complementary DNA. Blockage of primer extension by an alkylated RNA base results in the production of a complementary DNA fragment one nucleotide shorter than that arising from incorporation of a dideoxynucleotide opposite the same base. Calibration is in nucleotides from the ez1 5′ end. Unalkylated RNA (lane −) served as a negative control to identify primer extension products unrelated to alkylation. The degree of chemical modification can be difficult to assess at sites where it is no greater than the basal level of termination by reverse transcriptase on an unalkylated RNA template. CMCT did not react well with guanosine nucleotides, precluding a direct assessment of base pairing by those residues. (B) Summary of the alkylation data obtained for the entire rne 5′ UTR. (●) Heavy alkylation; (▴) moderate alkylation. At the few positions where there was a discrepancy between the in vivo and in vitro DMS alkylation data, the score reflects the reactivity of the nucleotides in vivo. The boundaries of structural domains are delineated by brackets, and the Shine-Dalgarno element and initiation codon are underlined. A caret marks the site of insertion of a cytidylate residue to create the BsrGI site (TGTACA) of pEZ1000. Arrows indicate the boundaries of 5′ UTR deletions made subsequently (see Fig. 4).