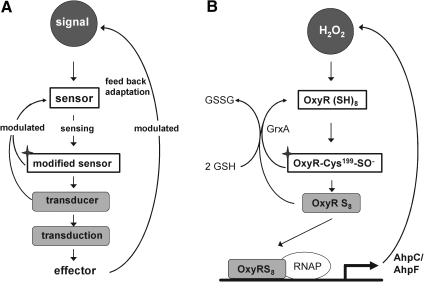
FIG. 1.
Scheme of transcriptional regulation and its implementation in bacterial redox control. (A) A minimalistic scheme of a regulatory circuit with the required elements: signaling molecule (black with white letters); sensor (white box); transducer(s) (light gray); and effector (typically the target gene). Termination reactions, which are subject to modulation, are indicated by reversed arrows. The labeling code is maintained in all following figures, if appropriate. (B) Demonstrates how this scheme is used in the OxyR regulation of prokaryotes. The signaling molecule is H2O2 targeting reduced OxyR as sensor. OxyR is first oxidized at Cys199 (with the oxidized sensor marked by a star) and stepwised further oxidized by H2O2. OxyR turns into a transducer (OxyRS8) and binds to its effector, the responsive element in the DNA, and allows the RNA polymerase (RNAP) to effect gene expression that removes the signal via enhanced alkyl hydroperoxide reductase (AhpC/AhpF) synthesis, thus terminating or modulating the sensing process. Prevention or termination of transduction is also achieved by reducing oxidized OxyR by glutaredoxin A (GrxA). The Grx system is modulated by glutathione (GSH) regeneration. In all eukaryotic systems additional transducers that are distinct from modified sensors are involved (see Fig. 2 and others).