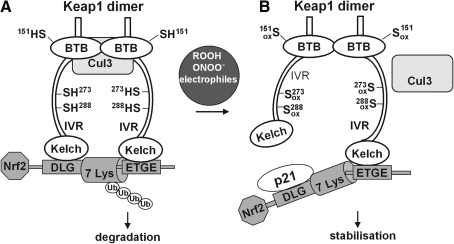
FIG. 7.
Model for redox-regulated Nrf2 activation based on conformational change of Keap1. In the resting state (A), Keap1 forms a homodimer via the broad complex/tramtrac/bric-a-brac (BTB) domains and associates to Nrf2 with its kelch domains. The Nrf2-ECH homology-2 (Neh2) domain of Nrf2 contains two Keap1 binding motifs with different affinity to the kelch domains, with ETGE representing the amino acid sequence with the higher affinity and DLG, the sequence with a much lower affinity. This two-site binding locks the central α-helix of the Neh2 domain of Nrf2 with is seven lysine residues (7 Lys) into a position suitable for ubiquitination. The intervening regions (IVR) contain the most reactive cysteine residues (Cys273 and 288) and build the adapter region for the Cullin-3 (Cul3)-based E3 ligase system. Nrf2 is polyubiquitinated and degraded by the proteasome. Under stressed conditions (B), oxidative/electrophilic modification of Cys272 and 288 induces a conformational change of Keap1 resulting in the dissociation from the weaker binding DLG motif in Nrf2, whereas binding to the ETGE motif remains [hinge and latch model as proposed by (484)]. P21 interacts with the DLG motif and supports dissociation of Nrf2 from this site. In contrast, modification of Cys151 in the BTB domain does not change the conformation of Keap1 but rather disrupts BTB Cul3 interaction. By both modifications ubiquitin ligase activity is lost and newly synthesized and released Nrf2 can move into the nucleus.