Figure 5.
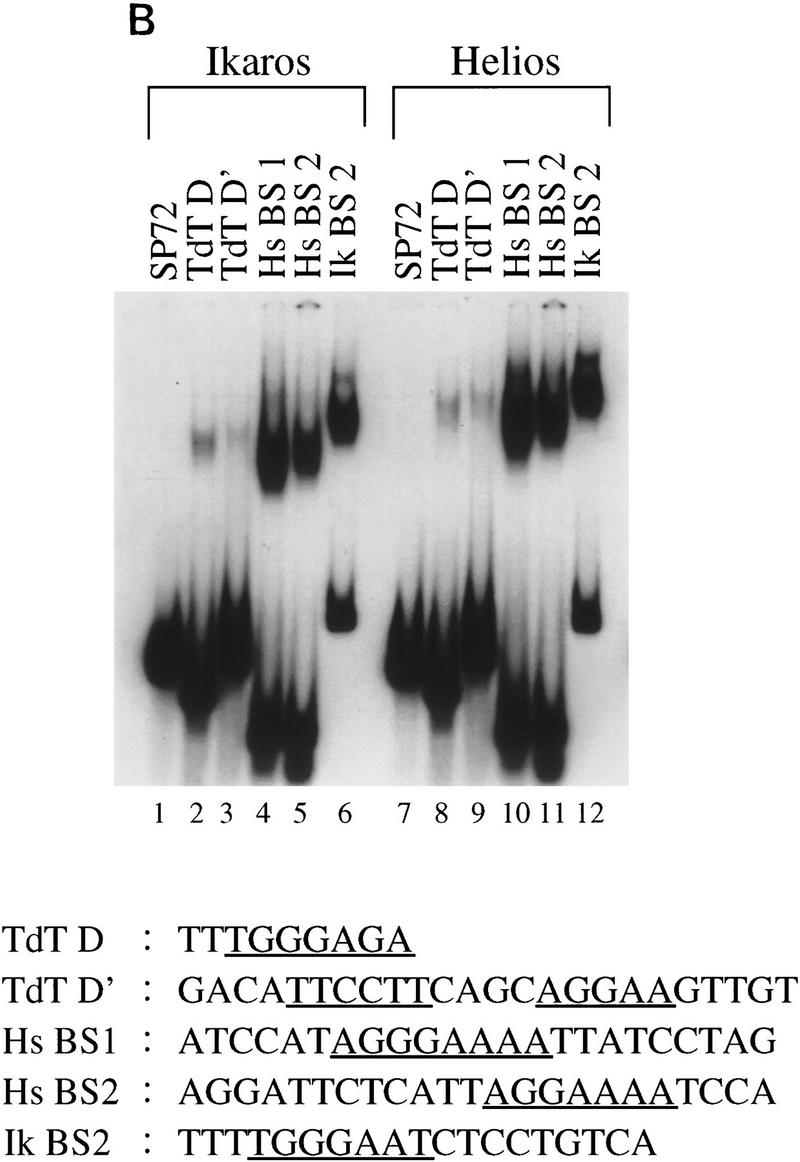
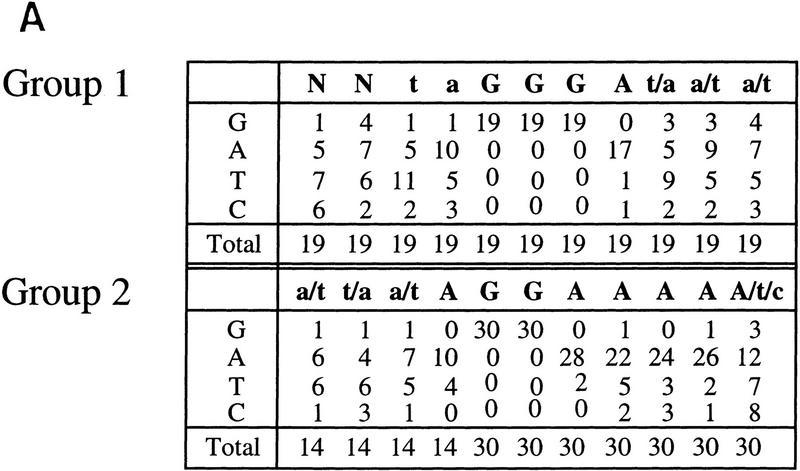
Helios A and Ikaros isoform V bind to similar DNA sequences. (A) High-affinity Helios A-binding sites were selected by a GST pull-down assay (Zweidler-McKay et al. 1996). DNA fragments selected by the GST–Helios A fusion protein were sequenced and tabulated. The selected sequences can be divided into two groups, both containing a core sequence of GGA. Group 1 was represented by 19 sequences and Group 2, by 30 sequences. The number of fragments containing each nucleotide at a given position following alignment are indicated for each group. Only 14 sequences are shown for the first four positions of the Group 2 sequence because the remaining 16 were derived from fragments in which these four positions were contained within the invariant primer-binding sites flanking the variable sequences. (B) Gel mobility shift assays were performed with recombinant GST–Ikaros isoform V (lanes 1–6) and recombinant Helios A (lanes 7–12) with probes containing the five sequences shown at the bottom (lanes 2–6, 8–12) and a negative control probe (lanes 1,7).The specific probes were derived from the pSP72 (Promega) multiple cloning site region (lanes 1,7), the TdT D (lanes 2,8) and D′ (lanes 3,9) elements, the Hs BS1 (i.e., Helios binding site 1; lanes 4,10) and Hs BS2 (lanes 5,11) sequences selected in this study, and the IkBS2 (lanes 6,12) sequence selected previously (Molnar and Georgopoulos 1994).