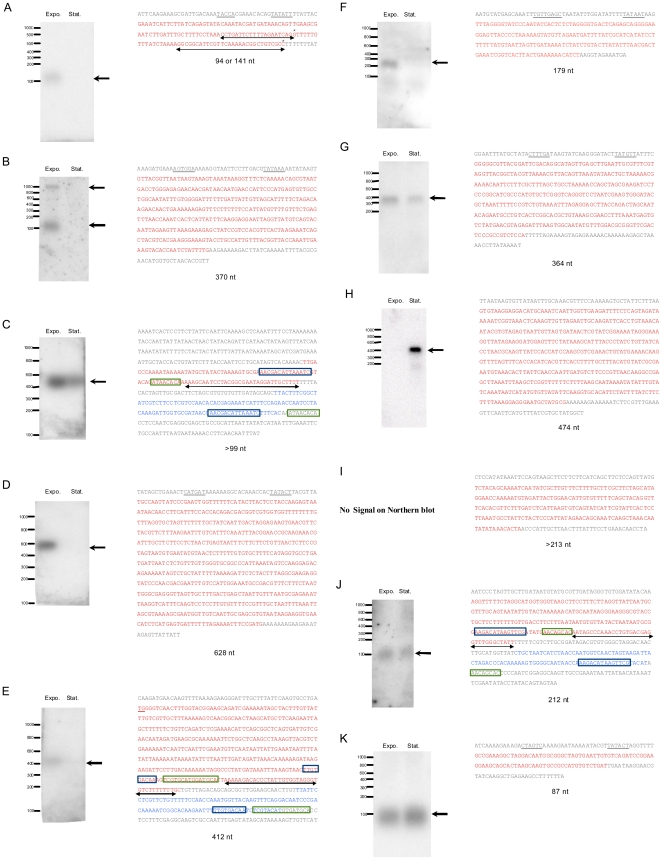
Figure 1. Northern blots and sequences of sRNAs (A: EF3314_EF335, B: EF0820_EF0821, C: EFA0080_EFA0081, D: EF1368_EF1369, E: EF0408_EF0409, F: EF0605_EF0606, G: EF1097_EF1098, H: EF0869_EF0870, I: EF0136_EF0137, J: EFB0062_EFB0063 and K: EF2205_EF2206).
RNA was isolated from cells at exponential (Expo) and stationary (Stat) phases. Northern blot analyses were performed using α32P-labelled probes. Arrows on Northern blot picture indicate the sRNAs corresponding bands. The transcriptional start sites and terminators of sRNAs were determined by 5′ RACE and 3′ RACE or by in silico analysis using TransTerm software. The putative −10 and/or −35 promoter sequences are underlined, and the sRNA sequence is written in red letters. Putative 3′-ends of EF3314_EF335 sRNA (panel A) is indicated by stars (*). The 3′-end of the sequenceof EF0136_EF0137 (panel I) mentioned here corresponds to the 3′-end of the tiling array probe. Black arrows in the sequence indicate the predicted terminators. The fst gene is written in blue letters and direct repeats “a” and “b” (DRa and DRb) of par system are blue and green boxed, respectively (panels C, E, and J).