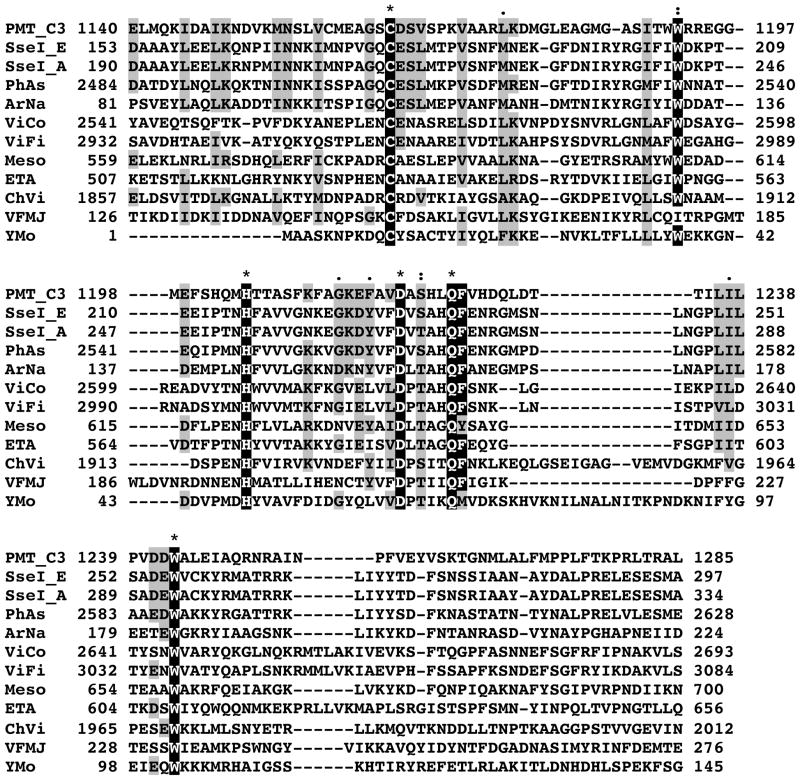
Figure 2. Alignment of amino acid sequences with similarity to PMT-C3.
The protein sequences were obtained from NCBI; PMT_C3: C3 domain of Pasteurella multocida toxin; SseI_E: SseI from Salmonella enterica serovar Enteritidis; SseI_A: SseI from Salmonella enterica serovar Arizonae; PhAs: insecticidal toxin from Photorhabdus asymbiotica; ArNa: secreted effector protein from Arsenophonus nasoniae; ViCo: hypothetical protein VIC_001387 from Vibrio coralliilyticus; ViFi: hypothetical protein VF_A1129 from Vibrio fischeri strain ES114; Meso: hypothetical protein Meso_3517 from Mesorhizobium sp. Strain BNC1; ETA: hypothetical protein ETA_29930 from Erwinia tasmaniensis strain Et1/99; ChVi: hypothetical protein CV_2593 from Chromobacterium violaceum; VFMJ: hypothetical protein VFMJ11_A0013 from Vibrio fischeri strain MJ11; YMo: hypothetical protein ymoll0001_35050 from Yersinia mollaretii. The numbers at the ends of each line correspond to the amino acid position in the indicated protein. The catalytic Cys-His-Asp triad as well as the highly conserved Trp and Gln-Phe residues are highlighted in black, “*” denotes identical amino acid residues; “:” denotes highly conserved residues; “.” denotes conserved residues.