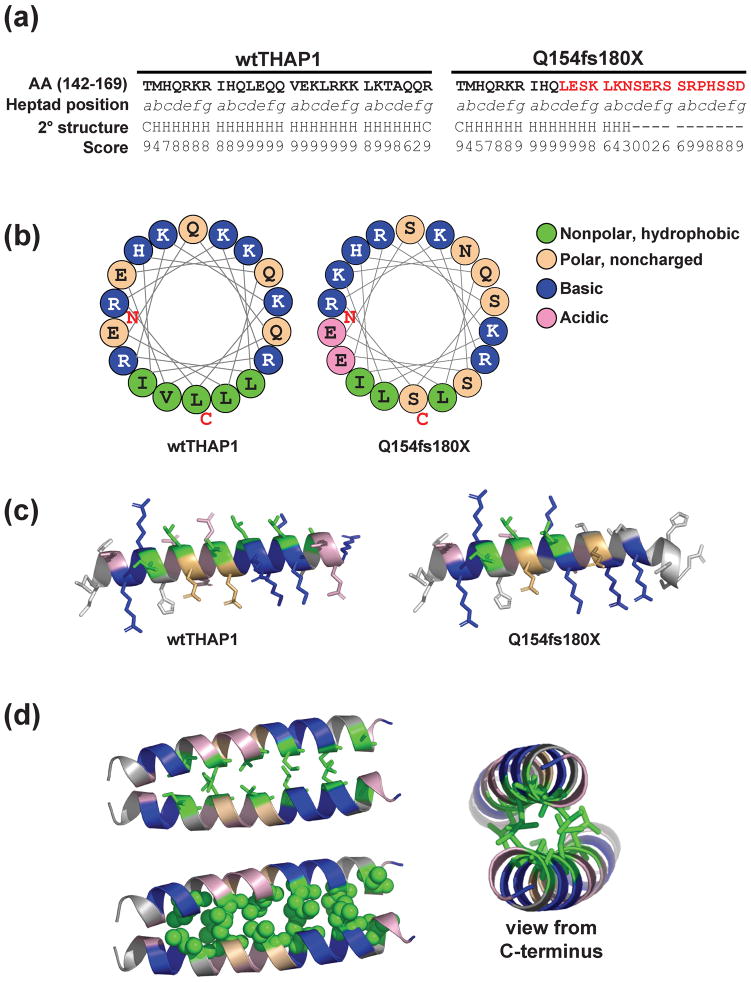
Fig. 6. Predicted structural model of THAP1 dimerization.
(A) Sequence of wtTHAP1 and the Q154fs180X corresponding to amino acids 142–169, representing the first four heptads of the predicted coiled-coil domain in wtTHAP1. Heptad position (abcdefg) is denoted below each residue, and the novel sequence encoded by the frameshift mutation is shown in red. Secondary structure analysis predicts a helical structure (H) for the wild-type sequence but not for most of the sequence encoded by the frameshift. Confidence scores for the secondary structure prediction determined by the PSIPRED Protein Structure Prediction Server are shown below each residue. (B) Helical wheel analysis of wtTHAP1 and Q154fs180X mutants. Examples shown represent residues 146–163. In wtTHAP1, clustering of hydrophobic residues on one face with basic/polar residues on the other suggests an amphipathic helix in this region. The Q154fs180X mutant loses this continuous stretch of hydrophobic residues due to polar serines which replace leucines at d3 and a4. (C) Richardson diagrams predicting the three-dimensional arrangement of hydrophobic (green), basic (blue), noncharged polar (tan), and acidic (pink) residues in this region. A potential hydrophobic interface is predicted in wtTHAP1 which is partially missing in Q154fs180X. (D) Predicted structural model of THAP1 dimerization at this potential hydrophobic interface. Model generated with Pymol software based on parallel coiled-coil domain of residues 164–191 of Protein Data Bank (PDB) 3Q0X. Side view of interacting monomers shown as stick (upper left) and space-filling (lower left) models. Alternate view from C-terminal (right) illustrates that basic arginine and lysine residues would be clustered on the outer, exposed faces of the predicted dimer structure.