Figure 2.
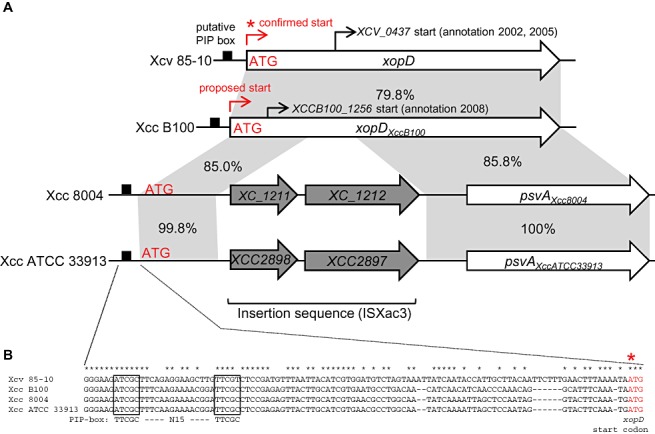
Comparison of the chromosomal regions spanning the xopD and xopD‐like loci in different Xanthomonas strains. (A) The promoter and open reading frame (ORF) for xopD from Xanthomonas campestris pv. vesicatoria strain 85‐10 (Xcv 85‐10), xopDXccB100 from X. campestris pv. campestris strain B100 (Xcc B100), psvAXcc8004 from X. campestris pv. campestris strain 8004 (Xcc 8004) and psvAXccATCC33913 from X. campestris pv. campestris strain ATCC 33913 (Xcc ATCC 33913). Black boxes, putative pathogen‐inducible promoter (PIP)‐boxes. Red asterisk, confirmed ATG start codon for the XopD protein. The proposed start site for xopDXccB100 is shown at the corresponding ATG denoted in red. The original start sites annotated for xopD (Noël et al., 2002; Thieme et al., 2005) and xopDXccB100 (Vorhölter et al., 2008) are noted by thin black arrows. The grey shading indicates identical DNA sequence shared between xopD and xopD‐like genes. The percentage sequence identity is noted for the adjacent highlighted regions. The xopD‐like loci in Xcc 8004 and Xcc ATCC 33913 are disrupted by an insertion sequence (ISXac3) containing two genes (i.e. XC_1211 and XC_1212 in Xcc 8004 and XCC2898 and XCC2897 in Xcc ATCC) resulting in a natural 5′ deletion, creating psvAXcc8004 and psvAXccATCC33913, respectively. (B) Sequence alignment of the xopD and xopD‐like promoter regions. Boxes indicate the putative PIP‐box sequences sharing identity with the consensus motif (TTCGC—N15—TTCGC). Red asterisk indicates the confirmed ATG region in the xopD gene.
