Figure 7.
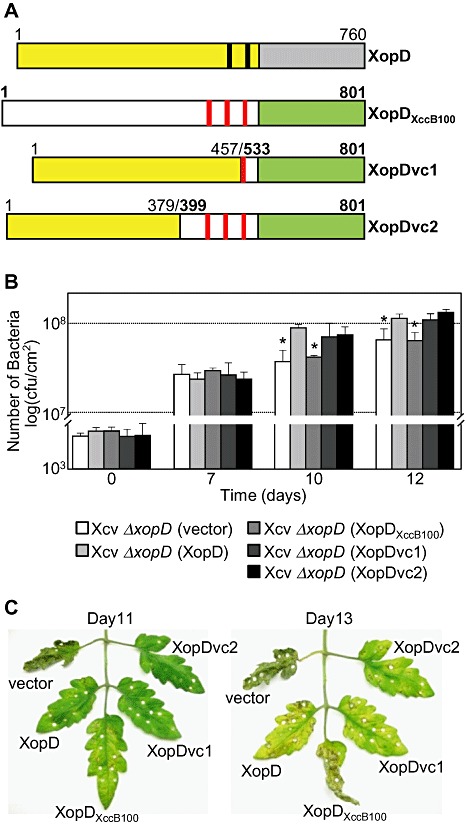
Chimeric XopD‐XopDXccB100 fusion proteins complemented Xanthomonas campestris pv. vesicatoria (Xcv) ΔxopD mutant phenotypes in infected tomato leaves. (A) Schematic diagram of wild‐type XopD (1–760 amino acids) and XopDXccB100 (1–801 amino acids) and two chimeric fusion proteins, XopDvc1 and XopDvc2. XopDvc1 contains amino acids 1–457 of XopD fused to amino acids 533–801 of XopDXccB100. XopDvc2 contains amino acids 1–379 of XopD fused to amino aids 399–801 of XopDXccB100. The yellow rectangles represent XopD's N‐terminal domain before its small ubiquitin‐related modifier (SUMO) protease domain (grey rectangle), and the black bars represent the putative ethylene‐responsive element binding factor‐associated amphiphilic repression (EAR) motifs. The white rectangles represent XopDXccB100's N‐terminal domain before its SUMO protease domain (green rectangle) and the red bars represent putative EAR motifs. (B) Growth of Xcv strains in tomato leaves. Leaves were hand‐inoculated with a 1 × 105 cells/mL suspension of Xcv ΔxopD (vector) containing pVSP61 (white bar), Xcv ΔxopD (XopD) containing pVSP61(lacZ promoter‐XopD‐His) (light grey bar), Xcv ΔxopD (XopDXccB100) containing pVSP61(lacZ promoter‐XopDXccB100‐His) (grey bar), Xcv ΔxopD (XopDvc1) containing pVSP61(lacZ promoter‐XopDvc1‐His) (dark grey bar) or Xcv ΔxopD (XopDvc2) containing pVSP61(lacZ promoter‐XopDvc2‐His) (black bar). Data points represent mean log10 colony‐forming units (cfu)/cm2± standard deviation (SD) of three tomato plants. Error bars indicate SD. The asterisks above the bars indicate statistically significant (t‐test, P < 0.05) differences between the bacterial numbers for Xcv (XopD) and Xcv ΔxopD (vector) or Xcv ΔxopD (XopDXccB100). (C) Phenotype of the Xcv‐infected tomato leaves sampled in (B). Hole punches were used for the quantification of the bacterial numbers depicted in (B). Leaves were photographed at 11 and 13 days post‐inoculation. Similar phenotypes were observed in three independent experiments.
