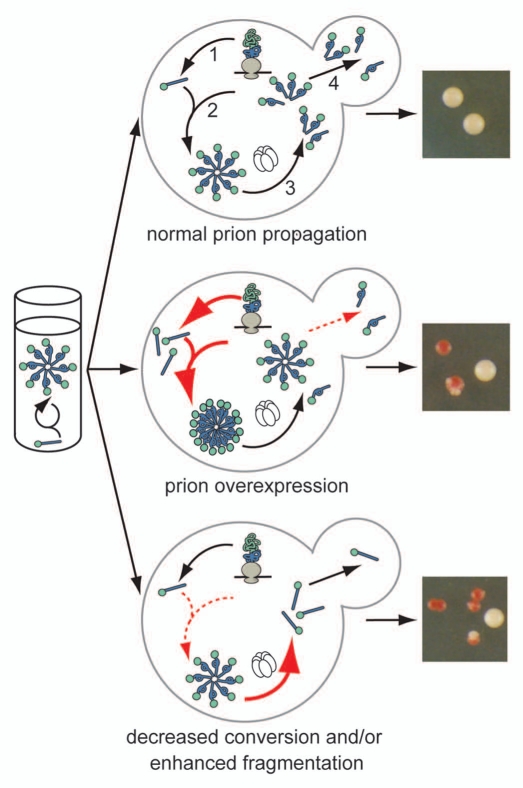
Figure 1.
The cellular environment modulates prion misfolding pathways to create protein-based traits. (top) Self-replicating protein conformations create stable phenotypes (white colonies) in vivo when the processes of synthesis (1), conversion (2), fragmentation (3) by Hsp104 (hexamer) and transmission (4) are balanced to allow aggregates of prion proteins to persist in vivo. (middle) Overexpression of a prion protein promotes the conversion reaction (red arrows), leading to the accumulation of large aggregates that are inefficiently transmitted to daughter cells (dotted red arrow) and loss of the prion-associated phenotype (red colonies). (bottom) Dominant inhibition of prion propagation by mutants that decrease conversion efficiency (dotted red arrow) or enhance fragmentation efficiency (solid red arrow) promote aggregate disassembly (ball and stick) and induce prion loss (red colonies).