Figure 3.
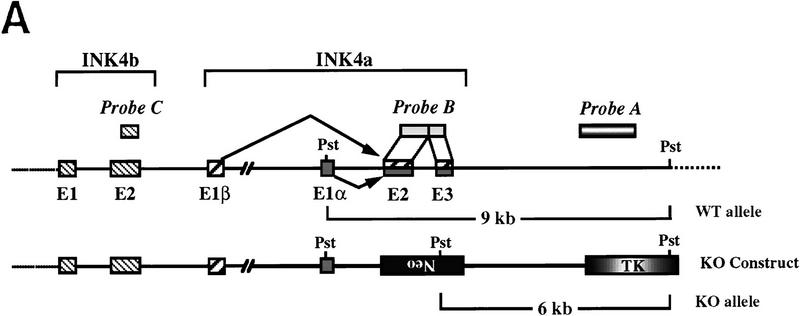
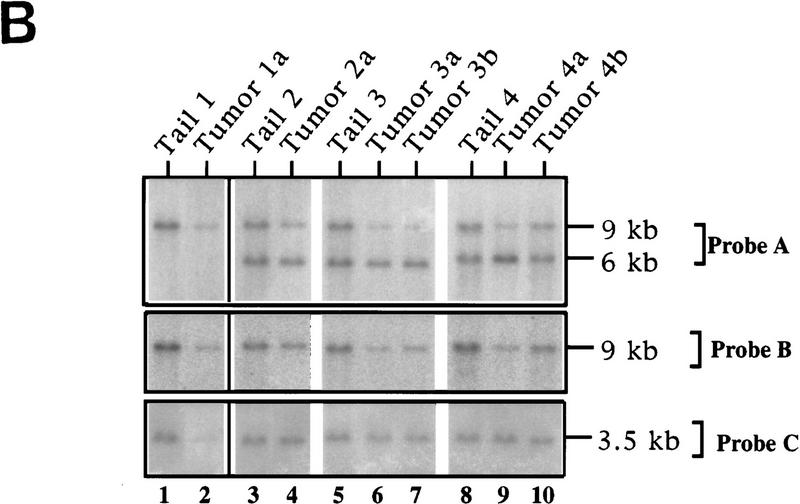
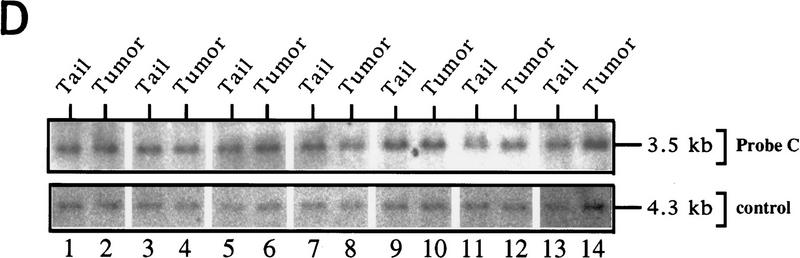
LOH of ink4a in ras-induced malignant melanoma. (A) Targeting strategy of ink4a locus. Restriction map of the wild-type and knockout ink4a alleles and probes used. (B) Southern blot analysis of tumor DNAs extracted from a panel of Ras-induced cutaneous melanomas and their respective normal DNAs extracted from tails. (Top) Hybridization to the 3′-flanking probe (probe A), which distinguishes the wild-type and knockout alleles. Note consistent reduction in hybridization signal from the remaining wild-type allele in tumor samples examined when compared with that in the corresponding tail DNA samples. The strength of the residual wild-type signals correlates well with the amount of normal tissue composition within the corresponding tumor. More importantly, the derivative cell line of tumor 4a (lane 9) reveals a complete loss of wild-type signal by Southern blot analysis (data not shown). The signal ratios of wild-type to knockout alleles, calculated as described (see Materials and Methods), are as follows: (Lanes 3,4) 1.0/0.76; (lanes 5–7) 1.0/0.63/0.43; (lanes 8–10) 1.0/0.40/0.83. (Middle) Hybridization to an internal probe (probe B) that corresponds to the deleted region in the knockout allele. (Bottom) Hybridization to exon 2 probe of ink4b (probe C). (C) Multiplex PCR analysis. (Top) Generation of standard curve; (left) autoradiograph of ink4a exon 2 signal; (right) linear regression performed on exon 2 signals, as determined by PhosphoImager quantitation, plotted against amount of wild-type DNA. (Bottom) Amplification of exon 2 signal and determination of reduction to homozygosity in tumor samples. (D) Southern blot analysis of normal and tumor DNAs from melanomas arising in ink4a−/− background. Loading is normalized against hybridization to control, an irrelevant single copy gene probe (see Materials and Methods). ink4b hybridization signal intensities, calculated as described (see Materials and Methods), are as followes: (Lanes 1,2) 1.0/1.2; (lanes 3,4) 1.0/1.0; (lanes 5,6) 1.0/1.1; (lanes 7,8) 1.0/0.87; (lanes 9,10) 1.0/0.84; (lanes 11,12) 1.0/1.2; (lanes 13,14) 1.0/1.0.