Fig. 1.
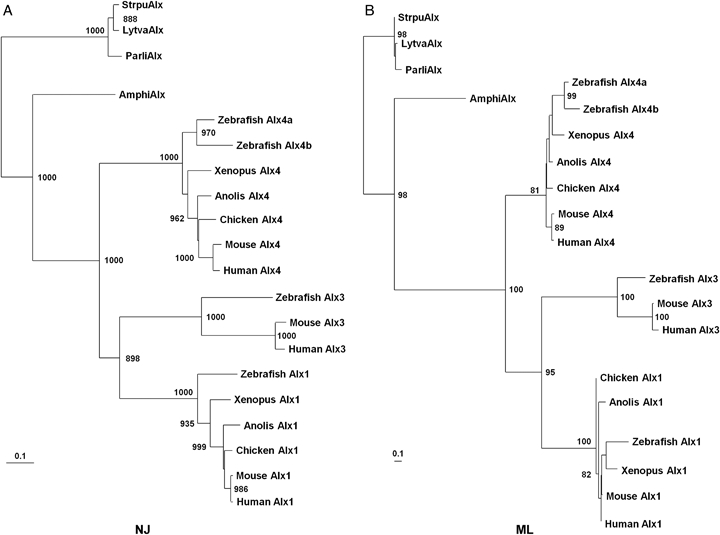
Molecular phylogenetic trees showing the relationships between deuterostome Alx proteins based on neighbor-joining (NJ, A) and maximum-likelihood (ML, B) analyses. Echinoderm Alx proteins were used as outgroups. Branch lengths are proportional to evolutionary distance corrected for multiple substitutions; the scale bar denotes 0.1 underlying amino acid substitutions per site. Figures on branches indicate robustness of each node (>70%), estimated from 1000 bootstrap replicates for NJ (A) and 100 replicates for ML (B). AmphiAlx (GenBank: JF460798) is clearly shown to be an ortholog of vertebrate Alx genes by both the NJ (100%) and ML (98%) methods. Furthermore, all extant bony vertebrates share at least two Alx gene duplication events: one that gave rise to the Alx4 and Alx1/3 paralogy groups (100% for both trees), and a second that gave rise to the Alx1 and Alx3 paralogy groups (89.8%, 95%). Further duplication of Alx4 genes is observed in zebrafish and other teleost fishes (see text).
