Figure 3.
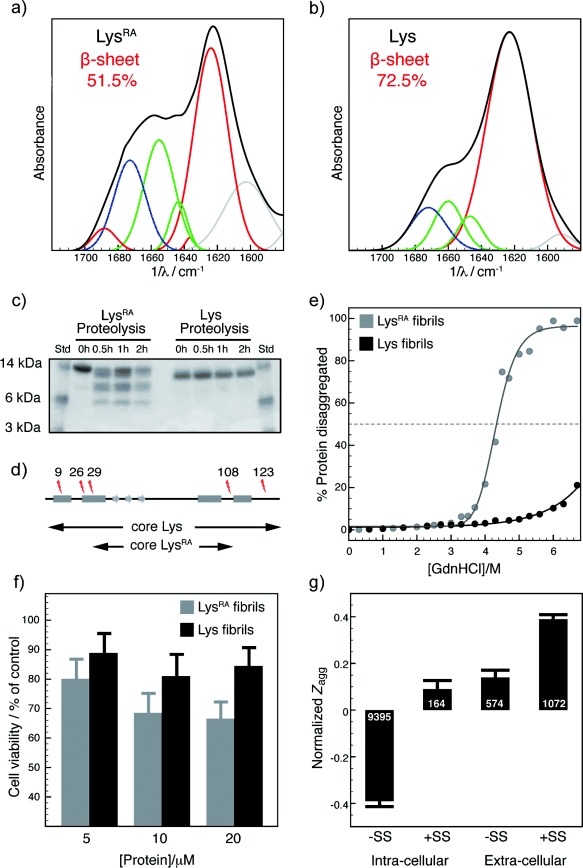
a, b) ATR-FTIR spectra of LysRA (a) and Lys fibrils (b), shown in black, with the contributions obtained by curve fitting colored as follows: red: β sheet, green: random/α helix, blue: turns and loops, gray: side chains. c) SDS-PAGE of fibril samples isolated by ultracentrifugation during proteolysis. d) Positions of cleavage sites indicated on the polypeptide chain; rectangles and arrows indicate the location of the native α helices and β strands. The protease-resistant region of LysRA fibrils encompasses residues 29–108; almost exactly the same region (32–108) was found to be protease-resistant in early aggregates of Lys,[24] but maturation progressively transforms these species into fibrils protected from proteolysis (E. Frare, personal communication). e) Disaggregation by GdnHCl of fibrils formed by LysRA (gray) and Lys (black). f) Effect of fibrils formed by LysRA (gray) and Lys (black) on SH-SY5Y cells evaluated by the calcein viability assay; the mean and the 95 % confidence interval after three experiments are reported. g) Normalized aggregation propensity of the human proteome as a function of cellular localization and of the presence of disulfide bonds. The numbers in white represent the number of sequences belonging to each class.
