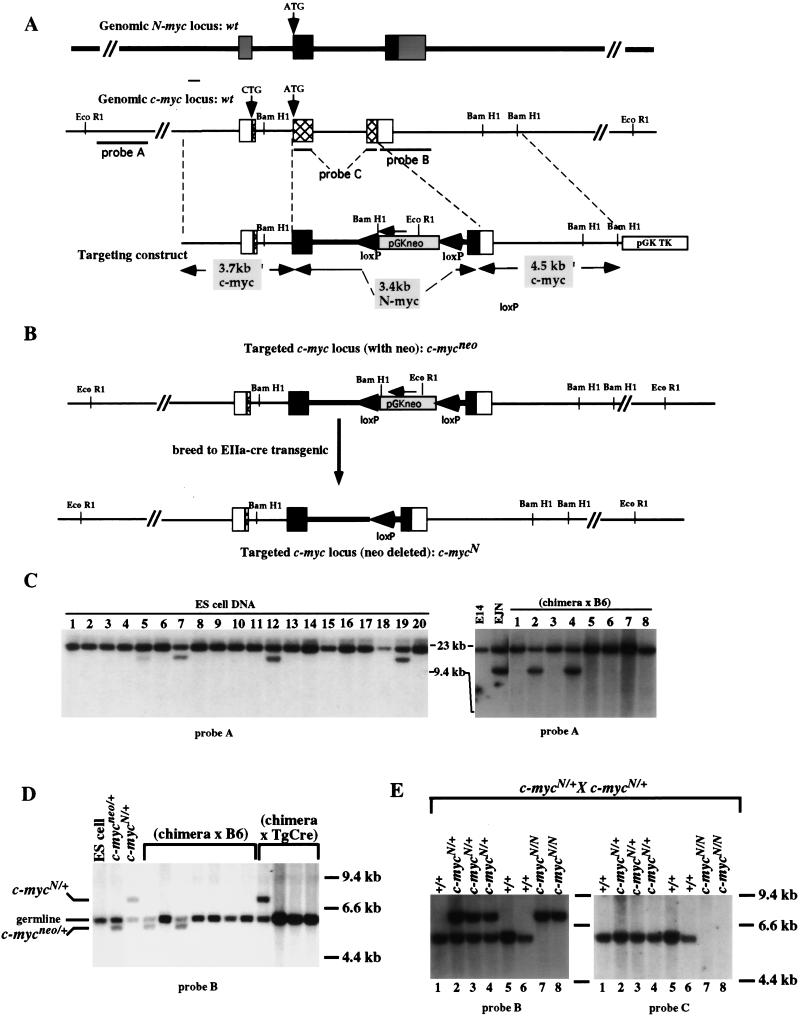
Figure 1.
Targeted replacement of c-myc coding sequences with N-myc coding sequences. (A) Schematic diagrams of the intron–exon structures of the genomic N-myc and c-myc loci and the N- into c-myc replacement construct. The majority of the coding sequences of N-myc (black boxes) and c-myc (hatched boxes) are contained within exons 2 and 3. The 5′ and 3′ untranslated regions are represented as shaded boxes for N-myc and open boxes for c-myc. In the targeting construct, called pNCR11, N-myc exons 2 and 3 and the intervening intron are flanked by 5′ and 3′ c-myc homology arms. PGKneo, surrounded by loxP sites, was inserted within the N-myc intron. (B) Schematic diagrams of the targeted c-myc loci before and after Cre-mediated recombination. pNCR11 was transfected into embryonic stem cells and screened for homologous recombination into the c-myc locus. A schematic of the structure of the targeted locus, called c-mycneo, is shown. Mice derived from the targeted ES cells were bred to transgenic mice carrying the EIIa–cre transgene and offspring were screened for transmission of the Cre-mediated recombination, resulting in deletion of the neo gene. A schematic of the structure of the targeted locus, called c-mycN, is shown. (C) Southern blot analysis for germ-line transmission of the c-mycneo targeted locus. Targeting of ES cells was screened by digestion of DNA with EcoRI and hybridization to probe A (left, lanes 5,7,12,19). Appropriate targeting was confirmed by digestion with multiple restriction enzymes and probes (data not shown). Targeted ES cells were injected into blastocysts and chimeric mice were bred either to C57BL/6 or 129Sv/Ev mice. Offspring were tested for germ-line transmission by digesting tail DNA samples with EcoRI and hybridizing to probe A (right, lanes 2,4; data not shown). (D) Southern blot analysis for Cre-mediated deletion of the neo gene to produce c-mycN/N mice. c-mycneo-transmitting chimera were bred to EIIa–cre transgenic mice. Tail-DNA samples were analyzed by digestion with BamHI and Southern blots were hybridized to probe B. The germ line BamHI fragment hybridizing to probe B is ∼6.0 kb; the c-mycneo allele results in a ∼5.5-kb fragment, because of inserted BamHI site in the PGKneo sequence. After Cre-mediated deletion of the PGKneo gene, the elimination of this BamHI site results in a probe B-hybridizing fragment of ∼7.0 kb. (E) Mating of heterozygous c-mycN/+ mice yielded homozygous mice that survived in the absence of a c-myc gene. Tail DNA from 10-day-old offspring of a c-mycN/+ heterozygous mating was digested with BamHI and Southern blots were hybridized to probe B (left). The diagnosed genotypes are designated above each lane and included wild type (c-myc+/+), heterozygous (c-mycN/+), and homozygous (c-mycN/N) (left). The Southern blot was stripped and hybridized to probe C. DNA from c-mycN/N mice failed to hybridize to this probe, which is composed of both coding exons of c-myc DNA, proving that these mice are unable to produce c-myc-encoded transcripts (right).