Figure 6.
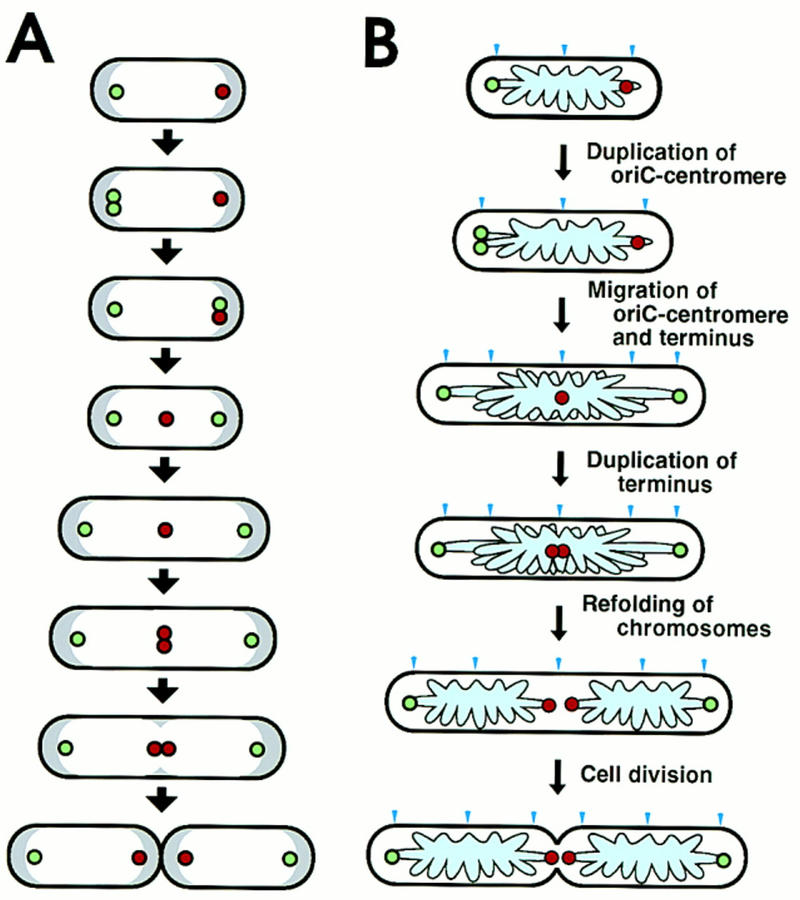
Schema of intracellular localization of replication origin and terminus during cell division cycle and a model of chromosome partioning in E. coli. (A) In cells growing with a 60-min doubling time, a replicated origin DNA segment (green circle) on the chromosome is localized near a nucleoid border in newborn cells. One copy of the replicated origin segment remains at the same position, whereas the other migrates to the opposite nucleoid border, where the replication terminus (red circle) is localized. Subsequently, the replication terminus migrates to midcell. The distance between an origin DNA segment and the nearest cell pole is constant during the cell division cycle, despite cell elongation. One round of chromosome replication is completed, and the terminus segment is duplicated at midcell. A septum is formed at midcell, resulting in newborn cells that have a replication terminus localized at the nucleoid border near a pole newly created by cell division. On the other hand, the replication origin is localized at the other nucleoid border, near the old pole. White and gray regions indicate nucleoid and cytosol spaces, respectively. (B) A model of partitioning of daughter chromosomes. A putative cis-acting sequence for migration, oriC–centromere, is located in a loop containing oriC. The specific migration patterns of oriC–centromere (green circle) and terminus (red circle) are shown in cells during the cell division cycle. The pale blue region and arrow head indicate the folded chromosome and the cell division site, respectively.
