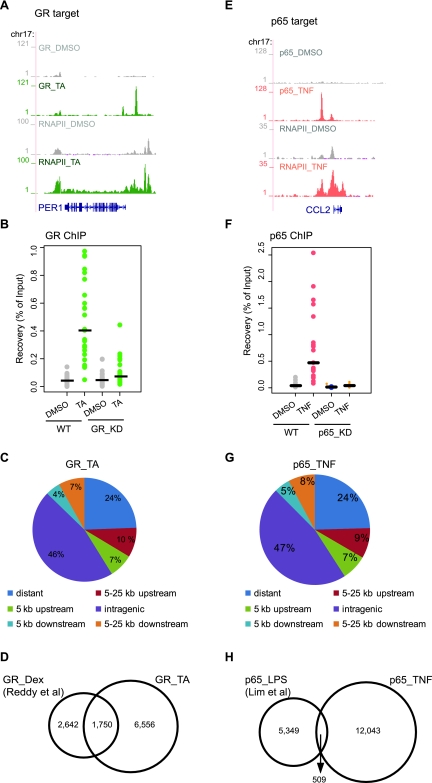
Figure 3.
Genome-wide GR- and p65-binding sites upon TA or TNF treatment. (A,E) RNAPII and GR (A) or p65 ChIP-seq data (E) illustrate RNAPII occupancy and GR and p65 binding in the promoter and/or within the gene body of target genes. Strong binding sites were detected upon TA (green) and TNF treatment (pink), whereas residual binding was observed in the absence of ligand (gray). Data were viewed in the University of California at Santa Cruz (UCSC) Genome Browser. The maximum number of overlapping tags, representing peak height, is indicated on the y-axis. (B,F) Validation of GR-binding sites (B) and p65-binding sites (F). Strip plots of GR ChIP–qPCR data for 25 randomly selected GR sites from WT and GR_KD cells (B) and p65 ChIP–qPCR data for 20 randomly selected p65 sites from WT and p65_KD cells (F) treated as indicated. Results are expressed as a percentage of input chromatin. Binding sites with fold induction (relative to DMSO) >2 are regarded as validated. Establishment of GR_KD and p65_KD cells are described in Supplemental Figure 2. (C,G) Genomic location analysis of GR-binding sites (C) and p65-binding sites (G). GR sites identified upon TA treatment (C) and p65 sites identified upon TNF treatment (G) were divided into the following categories based on their position relative to the nearest gene: distant (>25 Kb), within 5 Kb upstream, within 5 kb downstream, within 5–25 kb upstream, within 5–25 kb downstream and intragenic (exon, intron, and within 0–5 kb up- and downstream). The Pink Thing (http://pinkthing.cmbi.ru.nl) was used for the analysis. (D) Comparison of genome-wide GR profiling data. Venn diagram of the overlap of GR sites identified in the present study (8306 sites, ChIP-seq using HeLa B2 cells treated with TA for 4 h) with those identified in Reddy et al. (2009) (4392 sites, ChIP-seq using A549 cells treated with Dex for 1 h). (H) Comparison of genome-wide p65 profiling data. Venn diagram of the overlap of p65 sites identified in the present study (12,552 sites, ChIP-seq using HeLa B2 cells treated with TNF for 1 h) with those identified in Lim et al. (2007) (5858 sites, ChIP-PET using THP-1 cells treated with LPS for 1 h).