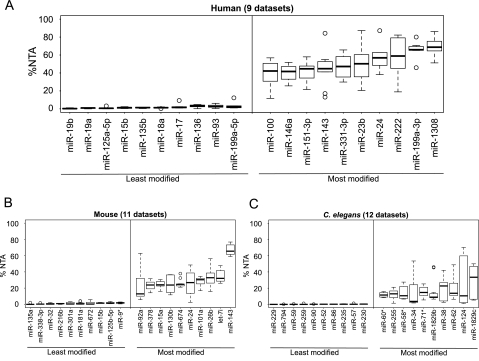
Figure 3.
MicroRNA-specific 3′ nontemplated nucleotide addition is seen across small RNA sequencing data sets. (A) A boxplot of the miRNAs demonstrating the lowest %NTA (i.e., least modified) and highest %NTA (i.e., most modified) across nine human sequencing data sets (data sets h1–h9 in Supplemental Table S1). MicroRNAs were required to have at least 40 reads in five or more of the nine data sets. (B,C) Boxplots of the miRNAs demonstrating the lowest %NTA (i.e., least modified) and highest %NTA (i.e., most modified) across 11 mouse cerebellum or medulloblastoma data sets (B) (data sets m1–m11 in Supplemental Table S1) and 12 C. elegans sequencing data sets (C) (data sets c1–c12 in Supplemental Table S1). MicroRNAs were required to have at least 40 reads in five or more of the data sets for mouse and for C. elegans to be included in the analysis.