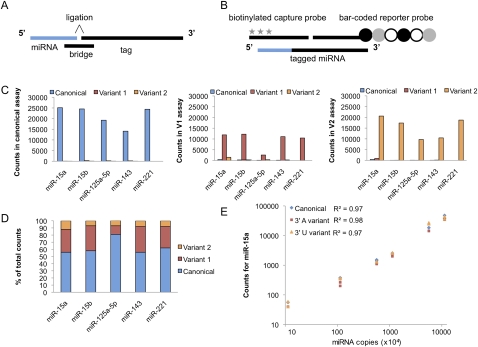
Figure 5.
Development of the nCounter miRNA assay to detect miRNA 3′ variants. (A) A schematic of the molecular components of the nCounter miRNA assay. The miRNA is shown in blue and DNA oligonucleotides in black. Each bridge oligonucleotide serves to template the 3′ end ligation of a particular miRNA species to a sequence-specific tag. Custom bridges were designed to discriminate between the 3′ end miRNA variants. (B) Following the removal of the excess tags and bridges, the tagged miRNAs are hybridized to specific capture and reporter probes attached to unique bar codes. The captured bar codes are individually resolved and counted in the NanoString nCounter assay system. The six-spot fluorescent bar code is represented by circles and the biotin capture moieties by stars. (C) Validation of the specificity of the nCounter assay. Three pools of synthetic RNA oligonucleotides, each containing canonical, variant 1, or variant 2 versions of five miRNAs were assayed. The graphs display the counts resulting when each of the three mixtures was individually assayed using the canonical (left), variant 1 (center), and variant 2 (right) bridge pools. (D) Validation of the accuracy of the nCounter assay. A mixture containing 60% canonical, 30% variant 1, and 10% variant 2 chemically synthesized miRNAs were assayed in each bridge pool. The relative abundance of each variant measured in the assay was then determined. (E) Validation of the linear range of the nCounter assays for miR-15a. Standard curves of synthetic oligonucleotides corresponding to the canonical, 3′ A (variant 1), or 3′ U (variant 2) form of miR-15a were assayed in their appropriate bridge pool. The graph displays the counts resulting from technical duplicates of standard curves ranging from 1 × 105 to 1 × 108 input copies of miRNA per reaction.