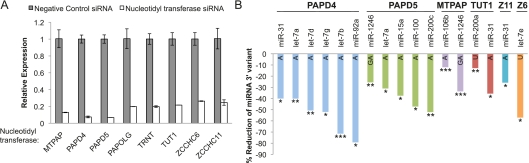
Figure 7.
MicroRNA 3′ nontemplated nucleotide additions are regulated by multiple nucleotidyl transferase enzymes. (A) qRT–PCR demonstrates the successful suppression of eight different nucleotidyl transferases in HCT-116 cells transfected, with siRNAs individually targeting each enzyme. Bars represent mean expression ±SD of biological replicates relative to cells transfected with siRNAs against the cyclophilin B negative control. Normalization was performed using qRT–PCR quantification of the endogenous control gene GUSB to control for variations in RNA input. (B) Bar graphs for each enzyme show percent reduction in miRNA 3′ variants following suppression of each of the six nucleotidyl transferases indicated above the x-axis. MicroRNAs shown represent variants that (1) were significantly reduced (FDR < 10%) in the cells treated with siRNAs targeting the enzyme of interest versus those treated with the negative control, siCyclophilin, and (2) showed at least a 5% absolute decrease in fractional abundance of the variant miRNA. The letter(s) within each bar for a given miRNA identifies which 3′ variant displays significant reduction. Z11 refers to the enzyme ZCCHC11 and Z6 to ZCCHC6. (*) miRNAs meeting criteria of FDR < 10%; (**) FDR < 5%; (***) FDR < 2%. The false discovery rate (FDR) is determined using the method of Benjamini and Hochberg. The change in fractional abundance and associated FDR for all canonical and 3′ miRNA variants retained after filtering for abundance is given in Supplemental Table S9.