An assembly process involving sequential recruitment of components and hierarchical dependency drives formation of the nuclear structures known as histone locus bodies.
Abstract
Nuclear bodies are protein- and RNA-containing structures that participate in a wide range of processes critical to genome function. Molecular self-organization is thought to drive nuclear body formation, but whether this occurs stochastically or via an ordered, hierarchical process is not fully understood. We addressed this question using RNAi and proteomic approaches in Drosophila melanogaster to identify and characterize novel components of the histone locus body (HLB), a nuclear body involved in the expression of replication-dependent histone genes. We identified the transcription elongation factor suppressor of Ty 6 (Spt6) and a homologue of mammalian nuclear protein of the ataxia telangiectasia–mutated locus that is encoded by the homeotic gene multisex combs (mxc) as novel HLB components. By combining genetic manipulation in both cell culture and embryos with cytological observations of Mxc, Spt6, and the known HLB components, FLICE-associated huge protein, Mute, U7 small nuclear ribonucleoprotein, and MPM-2 phosphoepitope, we demonstrated sequential recruitment and hierarchical dependency for localization of factors to HLBs during development, suggesting that ordered assembly can play a role in nuclear body formation.
Introduction
Nuclear organization is critical for genome function and the maintenance of genome integrity. Important to this organization are the macromolecular assemblies of protein- and RNA-containing particles, collectively called nuclear bodies, that participate in essential genome functions, including transcription, RNA processing, and DNA repair (Dundr and Misteli, 2010). The full composition of nuclear bodies, their mechanism of assembly, and how they contribute to nuclear processes, including gene expression, are not fully understood. Nuclear bodies are dynamic structures whose formation is governed by the self-organization of individual components (Misteli, 2001). However, a major unresolved question is whether self-organization occurs stochastically or via a hierarchical process in which the recruitment of certain components into the body depends on the prior recruitment of other components (Matera et al., 2009; Dundr and Misteli, 2010). Our ability to address this question for any particular nuclear body would benefit from the discovery of the complete suite of body components and a genetic system to mechanistically dissect how these components contribute to body assembly and function. We have begun to address this issue by examining the Drosophila melanogaster histone locus body (HLB), an evolutionarily conserved nuclear body associated with replication-dependent histone genes.
The bulk of histone protein synthesis is restricted to the S phase of the cell cycle by tightly coupling histone mRNA accumulation with DNA synthesis (Marzluff et al., 2008). This regulation is achieved through the action of cell cycle–regulated transcription and pre-mRNA–processing factors that produce histone mRNAs ending in an evolutionarily conserved stem loop rather than a poly(A) tail (Dominski and Marzluff, 2007). The unique histone mRNA 3′ end is formed by an endonucleolytic cleavage that requires both the stem loop and a sequence downstream of the cleavage site termed the histone downstream element. These cis-acting elements direct the recruitment of factors that stimulate pre-mRNA processing, including the U7 small nuclear RNP (snRNP), which binds directly to the histone downstream element, and the stem loop–binding protein (SLBP), which binds the stem loop (Dominski and Marzluff, 2007).
Cytological studies revealed that U7 snRNP accumulates in nuclear bodies that associate with replication-dependent histone genes. These bodies were first described in Xenopus laevis oocytes (Wu and Gall, 1993) and subsequently in mammalian cells (Frey and Matera, 1995). They were initially thought to be a subset of Cajal bodies (CBs), which are ubiquitous nuclear bodies that participate in snRNP assembly and that are identified by the presence of coilin protein (Handwerger and Gall, 2006; Matera et al., 2009). In Drosophila cells, foci of U7 snRNP also associate with histone genes, and in many tissues, these U7 snRNP foci are distinct from CBs (Liu et al., 2006, 2009). Consequently, the Drosophila U7 snRNP foci were termed HLBs to distinguish them from CBs (Liu et al., 2006). In mammalian cells, both nuclear protein of the ataxia telangiectasia–mutated locus (NPAT), a substrate of the G1-S regulatory kinase cyclin E/Cdk2 involved in histone gene expression (Ma et al., 2000; Zhao et al., 2000; Wei et al., 2003; Ye et al., 2003), and FLASH, a protein that interacts with U7 snRNP and is essential for histone pre-mRNA processing (Yang et al., 2009), are found in nuclear bodies lacking coilin that localize to histone genes (Bongiorno-Borbone et al., 2008; Ghule et al., 2008). Thus, mammalian cells contain nuclear bodies analogous to Drosophila HLBs.
The mechanism of HLB assembly and how HLBs function in histone mRNA biosynthesis are not known. To address these questions, we took advantage of a reagent that marks “active” HLBs during the S phase of Drosophila cells. The MPM-2 monoclonal antibody, which was raised against human mitotic phosphoproteins (Davis et al., 1983), labels nuclear foci in Drosophila cells when cyclin E/Cdk2 is active (Calvi et al., 1998). We recently demonstrated that these foci correspond to HLBs (White et al., 2007). We therefore used MPM-2 in two complementary approaches to identify novel HLB components. We first used mass spectrometry to identify proteins that immunoprecipitate with MPM-2 antibodies, either directly or indirectly. Because MPM-2 detects epitopes on many proteins, this approach could identify both HLB proteins and proteins that play no role in HLB assembly, regulation, or function. We therefore also developed an RNAi screen to identify genes required for the formation of MPM-2–positive HLBs. Together, these approaches identified two novel HLB components that, when analyzed in combination with previously identified HLB components, provide evidence for hierarchical self-organization during HLB formation.
Results
Identification of novel HLB components required for histone mRNA biosynthesis
A proteomic approach identifies suppressor of Ty 6 (Spt6) as an HLB protein.
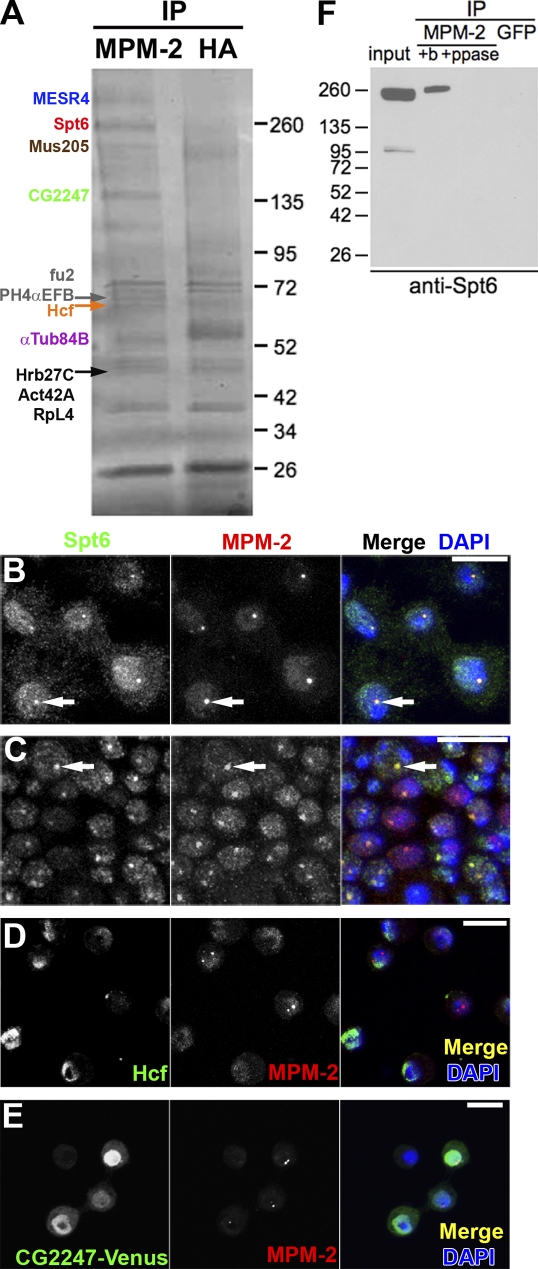
We used an immunoprecipitation (IP)/mass spectrometry approach as an initial strategy to identify MPM-2–reactive HLB proteins. Proteins that immunoprecipitated from S phase–arrested S2 cell nuclear extracts with MPM-2, but not with control anti-HA antibodies, were identified by mass spectrometry (Fig. 1 A). The MESR4 (misexpression suppressor of ras 4), Hcf (host cell factor), Spt6, and CG2247 proteins were unambiguously identified in three independent experiments. MESR4 has been implicated in RAS1 signaling, but its precise function is not known (Huang and Rubin, 2000). Hcf is part of the Drosophila ATAC (Ada2A containing) histone acetyltransferase complex that promotes nucleosome sliding by chromatin remodeling complexes (Guelman et al., 2006; Suganuma et al., 2008). Spt6 stimulates the movement of RNA polymerase II past nucleosomes (Andrulis et al., 2000; Kaplan et al., 2000; Ardehali et al., 2009). CG2247 has not been previously studied.
Figure 1.
Mass spectrometry identifies Spt6 as an HLB component. (A) Proteins precipitated with MPM-2 or control α-HA antibodies from S phase–arrested S2 cell nuclear extracts were separated by SDS-PAGE and stained with Coomassie. (B and C) S2 cells (B) and w1118 postblastoderm embryo (C) stained with α-Spt6, MPM-2, and DAPI. Spt6 forms nuclear foci that colocalize with MPM-2 (arrows). (D) S2 cells stained with α-Hcf, MPM-2, and DAPI. (E) S2 cells transiently transfected with CG2247-Venus and stained with α-GFP (green), MPM-2, and DAPI. Bars, 10 µm. (F) Nuclear extracts from S phase–arrested S2 cells were pretreated with buffer (+b) or λ phosphatase (+ppase) before immunoprecipitation (IP) with MPM-2 or control α-GFP antibodies and analyzed by Western blotting with α-Spt6. Molecular masses are given in kilodaltons.
We investigated whether Hcf, Spt6, and CG2247 localize to HLBs. All three proteins display nuclear localization (Fig. 1, B–E), but only Spt6 is present in nuclear foci that colocalize with MPM-2 foci (Fig. 1, B and C) and the U7 snRNP–specific proteins Lsm10 and Lsm11 (not depicted). Anti-Spt6 staining was also observed throughout the nucleus as described previously (Fig. 1, B and C; Kaplan et al., 2000). We confirmed that Spt6 is specifically immunoprecipitated by MPM-2 and that phosphatase treatment of the extract disrupts this interaction (Fig. 1 F). We cannot discern from this experiment whether Spt6 is directly recognized by the MPM-2 antibody, or is part of a complex recognized by the MPM-2 antibody. These data identify Spt6 as an HLB component, and the requirements for Spt6 localization to HLBs are explored in a subsequent section.
A genome-wide RNAi screen identifies HLB proteins, including the NPAT homologue multisex combs (Mxc).
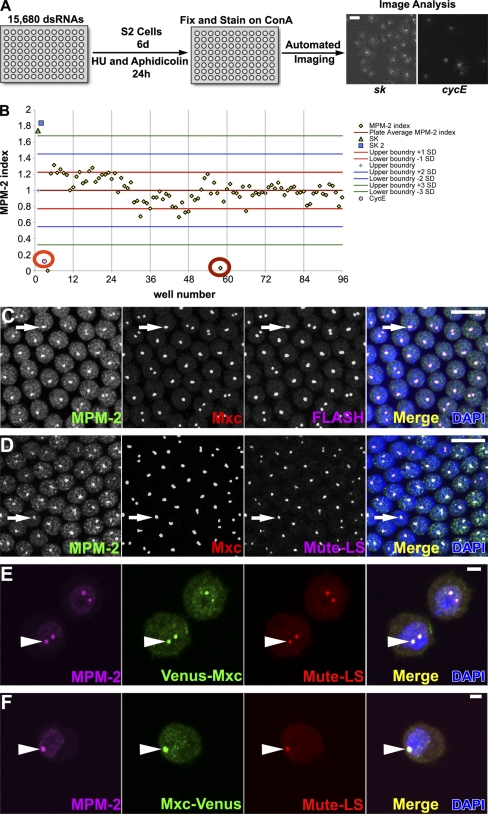
To identify additional HLB proteins, we developed a screen based on a high throughput microscopy assay capable of quantifying, in a population of synchronized Drosophila S2 cells, the percentage of nuclei containing at least one focus of MPM-2, a metric we designated the MPM-2 index. We hypothesized that double-stranded RNAs (dsRNAs) targeting an mRNA encoding (a) an MPM-2–reactive protein, (b) a protein required for the localization or phosphorylation of an MPM-2–reactive protein, or (c) a protein required for HLB formation would reduce the MPM-2 index.
To screen genome wide, S2 cells were transferred to 174 96-well plates containing a library of 15,680 dsRNAs targeting annotated Drosophila genes. The high throughput microscopy algorithm automatically determined MPM-2 indices by analyzing 50–400 S phase–arrested cells for each dsRNA treatment (Fig. 2 A). We identified 140 genes for which dsRNA treatment reduced the MPM-2 index to at least three standard deviations below the mean MPM-2 index of each plate (Fig. 2 B and Table S1). We then selected 95 genes from this original 140 to validate by rescreening in quadruplicate using a set of dsRNAs that target a different region of each gene than in the primary screen. To select these 95 genes, we eliminated many (e.g., those encoding ribosomal proteins) whose depletion is known to arrest the cell cycle outside of S phase (Table S1; Björklund et al., 2006). Because we expected the list of 95 genes to be enriched for potential positives, we used an MPM-2 index of 1.5 standard deviations below a control set of dsRNAs to verify positives. By this rescreening criteria, we identified eight genes that reduced the MPM-2 index in every replicate treatment (Table I, ++; and Table S2) and 18 genes that reduced the MPM-2 index by 1.5 standard deviations in at least one replicate (Table I, +; and Table S2).
Figure 2.
Genome-wide RNAi screen for MPM-2–negative HLBs. (A) Overview of the screen. Bar, 5 µm. (B) Graph of the MPM-2 indices from a representative screen plate. Note the mean MPM-2 index of the plate (cyan +), three standard deviations from this mean (green lines), the cyclin E (CycE)–positive control (orange circle), and a hit (red circle). (C and D) w1118 syncytial blastoderm (cycle 13) embryos stained with MPM-2, α-Mxc, DAPI, and α-FLASH or α–Mute-LS. Arrows indicate HLBs. Bars, 10 µm. (E and F) S2 cells expressing Venus-Mxc or Mxc-Venus with the Actin5C promoter stained with MPM-2 and α-GFP (green), which recognizes Venus, α–Mute-LS, and DAPI. Arrowheads indicate HLBs. Two HLB foci arise from unpaired homologous chromosomes. Bars, 2 µm. ConA, concanavalin A; HU, hydroxyurea; SK, pBluescript.
Table I.
Summary of HLB assembly and histone pre-mRNA processing requirements
| Gene transcript targeted by RNAi | Reduced MPM-2 index | Reduced Mxc index | Reduced FLASH index | Reduced Mute index | Scored positive with histone misprocessing reporter |
| mxc | ++ | ++ | ++ | ++ | Yesa |
| FLASH | ++ | + | ++ | + | Yes |
| Int6 | ++ | − | − | − | No |
| CSN3 | ++ | − | − | − | No |
| CSN4 | ++ | − | − | − | No |
| CSN5 | ++ | − | − | − | No |
| CG4849 | ++ | − | − | − | No |
| CG9769 | ++ | − | − | ++ | No |
| CG7597 | + | − | − | ++ | No |
| Tbp-1 | + | − | − | − | No |
| HLH106 | + | − | − | − | No |
| RpII18 | + | − | − | − | No |
| eIF-3p66 | + | − | − | − | No |
| alien | + | − | − | − | No |
| CG17841 | + | − | − | − | No |
| foi | + | − | − | − | No |
| Fer1HCH | + | − | − | − | No |
| CG11985 | + | − | − | − | No |
| CG18591/SmE | + | − | + | − | Yesa |
| MBD-R2 | + | − | ++ | + | Yes |
| CG18787/18789 | + | − | − | − | No |
| CG9121 | + | − | − | − | No |
| CG5844 | + | − | − | − | No |
| CG5147 | + | − | − | − | No |
| mfrn | + | − | − | + | No |
| CG14111 | + | − | − | − | No |
| Mute | − | − | − | ++ | No |
| Nnp-1 | − | − | − | + | No |
| raptor | − | − | − | + | No |
| MCRS1 | − | − | − | − | Yes |
| CG31111 | − | − | − | − | Yesa |
| CG8142 | − | − | − | − | Yesa |
| DMAP1 | − | − | − | − | Yesa |
| Dgt1 | − | − | − | − | Yesa |
| CG9772/Skp2 | − | − | − | − | Yesa |
The table lists those genes of the 95 selected for validation from the primary screen that scored in at least one of the five secondary assays: anti-MPM-2, -Mxc, -FLASH, or -Mute HLB labeling and activation of the histone pre-mRNA misprocessing reporter. A score of a double plus sign, a single plus sign, or a negative sign was assigned when the spot index fell ≥1.5 standard deviations from the control mean (i.e., −1.5 z score) in each, at least one, or no validation replicate, respectively (also see Table S2). Note that Mxc, FLASH, and Mute all strongly reduced their own index, indicating successful RNAi-mediated depletion. Although the eight genes listed at the bottom of the table did not produce a −1.5 or less z score in any of the MPM-2 validation replicates, they were included because they either activated the misprocessing reporter or affected the Mute index.
Newly identified genes involved in histone pre-mRNA processing.
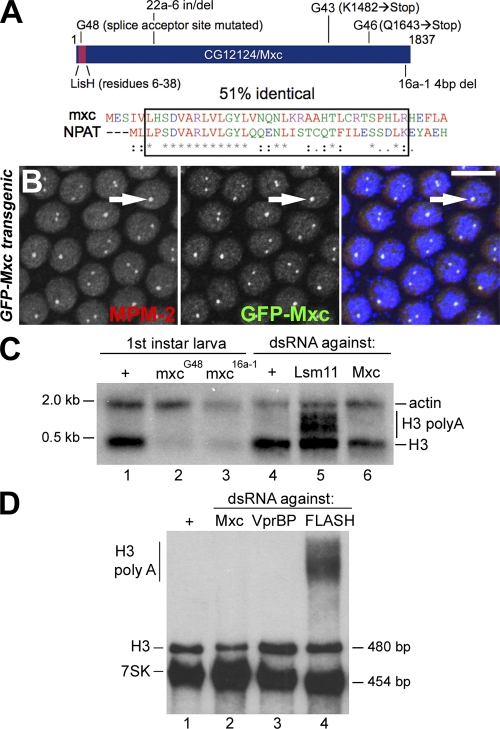
The RNAi screen should identify proteins (either resident in HLBs or not) required for HLB assembly and/or for production of the MPM-2 epitope. Because our primary goal was to identify HLB proteins, we focused on proteins known to localize to Drosophila HLBs or that are homologous to proteins known to localize to mammalian HLBs. CG12124 repeatedly scored strongly in our screens and is the previously described gene called mxc. mxc encodes a protein of 1,837 amino acids with an N-terminal lissencephaly homology (LisH) domain that is 51% identical to the LisH domain of human NPAT (see Fig. 5 A). Although the overall identity between Mxc and NPAT is only 12–15%, the NPAT LisH domain is more similar to the Mxc LisH domain than to any other LisH domain in the Drosophila genome. We therefore hypothesized that Mxc might be the Drosophila equivalent of mammalian NPAT.
Figure 5.
Mxc is essential for histone gene expression. (A, top) Diagram of CG12124/Mxc showing the position of the LisH motif and mutations G48 (AG to AA at the first intron/second exon border), 22a-6 (GTGTCAGCTG insertion for AA at N480), G43 (K1482→Stop), G46 (Q1643→Stop), and 16a-1 (TTCG 4-bp frame-shifting deletion [4bp del] at F1823 changing 5′-GAGTTCGAGGACATC-3′ to 5′-GAGAGGACATC-3′). (bottom) ClustalW alignment of the LisH domain from Mxc and NPAT. Red, green, blue, and purple letters indicate hydrophobic, hydrophilic, acidic, and basic amino acids, respectively. The box indicates the LisH domain. The asterisks and dots indicate identical and conserved amino acids, respectively. Single dots are used to show positions of similarity; double dots are used to show positions of high conservation. (B) Syncytial blastoderm (cycle 12) embryos expressing GFP-Mxc stained with α-GFP, MPM-2, and DAPI (blue). Bar, 10 µm. (C and D) H3 Northern analysis of the indicated genotypes or 7-d dsRNA treatments. A plus sign indicates wild-type larva (C, lane 1) or mock RNAi (C and D, lane 1). VprBP is a nonspecific control. Actin (C) and 7SK (D) were used as loading controls. There is 1.5 times as much RNA loaded in D, lane 2 than lanes 1, 3, and 4.
To determine whether Mxc localizes to HLBs, we raised specific antibodies to the C-terminal 169 amino acids of Mxc and also expressed N- or C-terminal Venus-tagged Mxc in S2 cells. Mxc localizes to HLBs in the early embryo (Fig. 2, C and D), and both forms of Venus-tagged Mxc localize to HLBs in S2 cells (Fig. 2, E and F). Mxc colocalizes with the known HLB proteins FLASH and muscle wasted (mute; Fig. 2, C and D), which were both also identified in our RNAi screen (Table S1). In both human and Drosophila cells, FLASH localizes to HLBs (Fig. 2 C; Barcaroli et al., 2006a,b; Yang et al., 2009). Mute was recently discovered in a screen for genes required for embryonic muscle development (Bulchand et al., 2010). The mute locus encodes long (Mute-L) and short (Mute-S) proteins produced from separate transcripts, and both Mute isoforms localize to HLBs (Fig. 2, D–F; Bulchand et al., 2010). The molecular function of the Mute protein is unknown. Thus, our genome-wide RNAi screen identified HLB proteins, including the novel component Mxc. The RNAi approach did not identify other known HLB components (e.g., U7 snRNP and Symplekin; Wagner et al., 2007), suggesting that these are not critical for HLB integrity. In addition, the HLB proteins identified by RNAi were not identified by IP/mass spectroscopy, likely because of their relatively low abundance in cells.
Mxc is MPM-2 reactive.
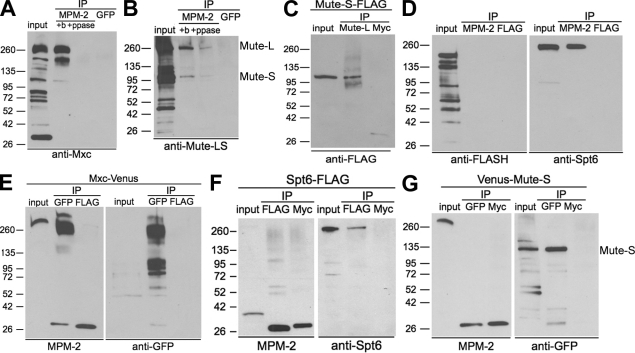
We hypothesized that MPM-2 might directly recognize one of the HLB proteins identified by our approaches (i.e., Spt6, FLASH, Mute, and Mxc). Although Mute and Mxc were not identified in the IP/mass spectrometry experiments, MPM-2 antibodies precipitate Mxc and both forms of Mute protein from S phase–arrested S2 nuclear extracts (Fig. 3, A and B). Mute-S is also precipitated by antibodies that react only with Mute-L, indicating that these isoforms are part of a complex (Fig. 3 C). In contrast, we did not detect FLASH in MPM-2 IPs, although we confirmed the presence of Spt6 (Fig. 3 D). This result indicates that FLASH is not an MPM-2–reactive protein. Pretreatment of nuclear extracts with λ phosphatase greatly reduces the amounts of Mxc, Mute, and Spt6 precipitated by MPM-2, indicating that the interaction between these proteins and MPM-2 antibodies depends on phosphorylation (Fig. 2 F and Fig. 3, A and B).
Figure 3.
Mxc is directly recognized by MPM-2. (A and B) Nuclear extracts from S phase–arrested S2 cells were pretreated with buffer only (+b) or λ phosphatase (+ppase) before immunoprecipitation (IP) with MPM-2 or GFP control antibodies and analyzed by Western blotting with α-Mxc (A) or α–Mute-LS (detects both the long and short isoforms of Mute; B). (C) α–Mute-L and α-Myc control IPs from cells expressing Mute-S–FLAG blotted with α-FLAG. (D) MPM-2 and α-FLAG control IPs from an S-phase–arrested S2 cell nuclear extract blotted with α-FLASH or α-Spt6. (E) α-GFP and α-FLAG control IPs from cells expressing Mxc-Venus blotted with MPM-2 or α-GFP. (F) α-FLAG and α-Myc control IPs from cells expressing Spt6-FLAG were blotted with α-Spt6 or MPM-2. (G) α-GFP and α-Myc control IPs from cells expressing Venus–Mute-S blotted with MPM-2 or α-GFP. Molecular masses are given in kilodaltons.
To test whether Mxc, Mute, and Spt6 are directly recognized by MPM-2, each protein was immunoprecipitated from S phase–arrested S2 nuclear extracts and analyzed by Western blotting with MPM-2 antibodies. Immunoprecipitated Mxc-Venus is reactive with MPM-2 on Western blots (Fig. 3 E). Note that there was some proteolysis of Mxc during the experiment. Because the Venus tag is on the C terminus, all the immunoprecipitated fragments must contain the C terminus of the protein. Only the high molecular weight fragments of Mxc react with MPM-2, suggesting that the phosphoepitope is in the N-terminal half of the protein. In contrast, immunoprecipitated Spt6-FLAG did not react with MPM-2 on Western blots (Fig. 3 F). Similarly, we do not detect any MPM-2 reactivity of Venus–Mute-S (Fig. 3 G). These data indicate that the HLB protein Mxc contains an MPM-2 epitope and suggest that Spt6 and Mute-S do not. Spt6 and Mute-S may precipitate with MPM-2 because they interact with phosphorylated Mxc and/or another MPM-2–reactive HLB protein. Because MPM-2 recognizes a cyclin E/Cdk2-dependent epitope in Drosophila HLBs, Mxc may be a cyclin E/Cdk2 substrate, as is human NPAT (Zhao et al., 1998; Ma et al., 2000).
Mxc is required for histone mRNA biosynthesis.
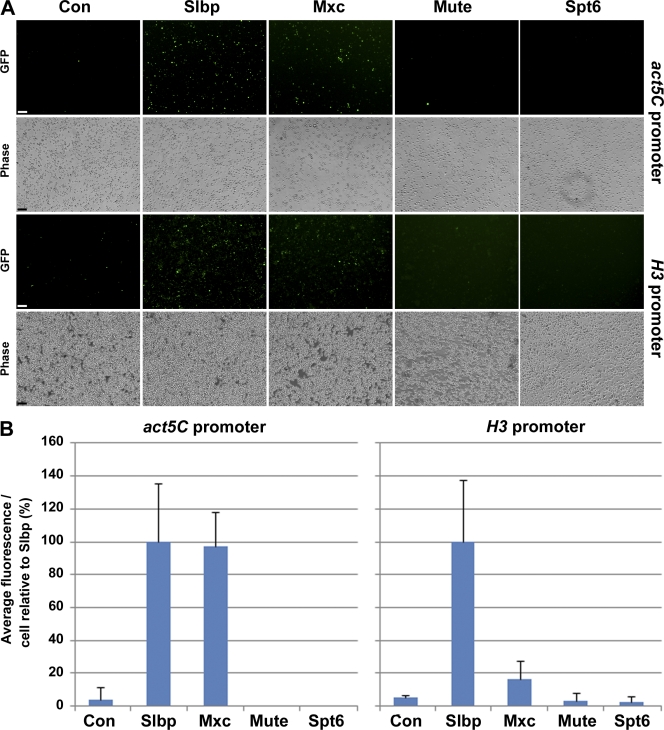
NPAT interacts with proteins necessary for histone H4 and H2b transcription and is thought to be a regulator of histone gene expression (Ye et al., 2003; Zheng et al., 2003; Miele et al., 2005; DeRan et al., 2008). To explore whether Mxc participates in histone gene transcription or pre-mRNA processing, we used two different reporter constructs that contain a histone 3′ processing signal located upstream of a GFP open reading frame (Fig. S1). These reporters result in GFP expression only when histone pre-mRNA processing is compromised. Each reporter utilizes a different promoter: one is driven by the act5C promoter (Yang et al., 2009), and the other is driven by the histone H3 promoter (Wagner et al., 2007). Note that if a factor is involved in both histone gene transcription and histone pre-mRNA processing, it will only score strongly with the reporter driven by the act5C promoter.
We compared Mxc depletion to the effect of depleting SLBP, which only affects histone pre-mRNA processing (Sullivan et al., 2001). Depletion of SLBP resulted in robust activation of both reporters (Fig. 4), which is consistent with SLBP playing a role in histone pre-mRNA processing and not in histone gene transcription. Depletion of Mxc activated the act5C-driven reporter but did not appreciably activate the H3-driven reporter, suggesting that loss of Mxc affected H3 promoter activity (Fig. 4). Thus, Mxc is possibly involved in both histone gene transcription and histone pre-mRNA processing. In contrast, depletion of Spt6 or Mute had no effect on either misprocessing reporter, suggesting that they are not involved in pre-mRNA processing (Fig. 4).
Figure 4.
GFP histone mRNA misprocessing reporter analysis. Dmel-2 cells stably transfected with the indicated GFP histone mRNA misprocessing reporter were treated for 5 d with dsRNAs directed against control (Con; PTB), SLBP, Mxc, Mute, or Spt6 dsRNAs. (A) Representative epifluorescence and bright-field images. Bars, 60 µm. (B) Quantification of GFP fluorescence/cell for each experiment. SLBP was set as 100 for each reporter. Error bars indicate standard deviations. Between 200 and 800 cells were quantified per experiment.
If Mxc were required for histone mRNA biosynthesis, mxc mutations would be expected to result in severe cell and developmental defects in vivo. The mxc locus on the X chromosome was originally defined by an allelic series of EMS and x ray–induced mutations that cause a variety of phenotypes depending on allele strength. These phenotypes include lethality, poor cell proliferation, and homeotic transformations that mimic the ectopic gain of function of BX-C and ANT-C genes (Santamaría and Randsholt, 1995; Saget et al., 1998; Remillieux-Leschelle et al., 2002). A recent entry on FlyBase by N. Randsholt (Centre National de la Recherche Scientifique, Paris, France) reported that mxc mutations are alleles of the gene originally annotated as CG12124. To directly test whether mxc mutations disrupt the function of CG12124, we generated transgenic flies expressing a GFP-Mxc fusion protein under the control of the constitutive ubiquitin promoter. GFP-Mxc localizes to embryonic HLBs and rescues the lethality caused by the mxcG48 allele (Fig. 5, A and B). We identified mutations predicted to disrupt CG12124 protein function by sequencing the mxcG48, mxcG46, mxc22a-6, mxc16a-1, and mxcG43 mutants (Fig. 5 A). S. Landais and L. Jones have obtained identical results for mxcG48, mxc22a-6, and mxcG43 (personal communication). mxcG48 is reported to be genetically null or nearly null (Santamaría and Randsholt, 1995; Saget et al., 1998), and our sequence data indicating a splice site acceptor mutation in exon 2 is consistent with this interpretation (Fig. 5 A).
To assess what roles Mxc might play in histone mRNA biosynthesis in vivo, we examined total H3 mRNA accumulation in mxcG48 mutants and in S2 cells treated with Mxc dsRNA. In both situations, we observe a reduction in total histone H3 mRNA but no detectable misprocessed H3 mRNA (Fig. 5, C and D). In contrast, knockdown of Lsm11 or FLASH results in substantial accumulation of misprocessed polyadenylated histone mRNA (Fig. 5, C and D). Thus, mutation of mxc results in failure to accumulate histone mRNA, which is consistent with a role for Mxc in histone gene expression. Whether this molecular phenotype is a cause or a consequence of the poor proliferation of mxc mutant cells cannot be discerned.
The RNAi screen identifies factors involved in histone pre-mRNA processing.
We also used S2 cells to ask whether the 95 hits selected from the primary MPM-2 screen play a role in histone pre-mRNA processing. Cells containing a stably integrated histone mRNA misprocessing reporter driven by the Act5C promoter (Fig. S1 B) were treated for 5 d with two independent aliquots from our library of 95 dsRNAs, and the amount of GFP/cell was quantified by fluorescence microscopy (see Materials and methods). 10 of the 95 dsRNAs significantly activated the reporter, scoring at least at the level of 5% of FLASH (Table I, column 6), which scores the strongest of any factor in this assay (Fig. S1 B; Yang et al., 2009). Of these 10, two (MBD-R2 and MCRS1) were identified in our previous genome-wide RNAi screen for histone pre-mRNA processing factors (Wagner et al., 2007). Seven, including Mxc, DMAP1, and Dgt1, were not previously identified as processing factors (Table I, footnote). Dgt1 is part of a histone acetyltransferase complex that contains MBD-R2 and MCRS1 (Mendjan et al., 2006; Cai et al., 2010; Prestel et al., 2010). Of the remaining four, two (CG31111 and CG8142) have not been previously characterized: CG9772 is an F-box protein homologous to Skp2 (Shibutani et al., 2008), and CG18591 is SmE, which is found in both spliceosomal snRNPs and U7 snRNP (Pillai et al., 2001, 2003). Inefficient knockdown of SmE likely accounts for SmE scoring in both the MPM-2 and misprocessing assays because efficient knockdown is cell lethal (unpublished data).
Developmental and genetic evidence in support of hierarchical HLB assembly
Mxc and FLASH are required for HLB assembly.
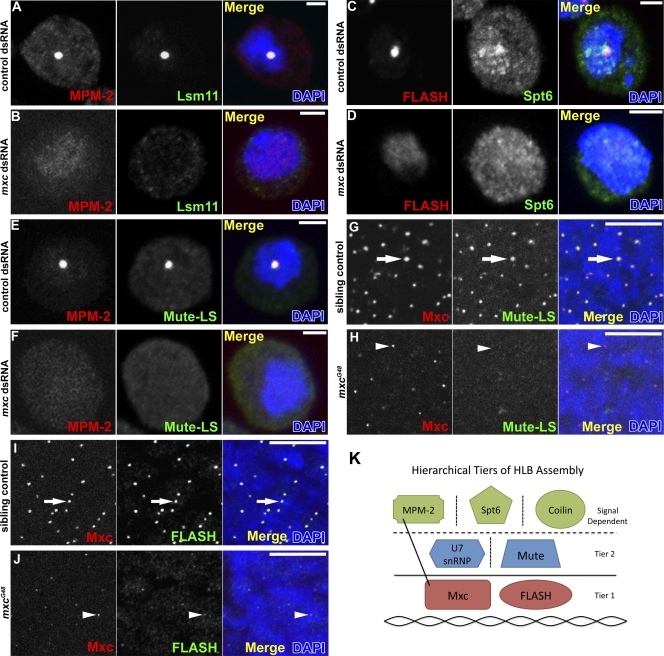
We also used antibodies to Mxc, FLASH, and Mute to assess HLB formation in S2 cells after dsRNA depletion of each of the 95 top hits from the primary MPM-2 screen (Table I and Table S1). For each dsRNA treatment, we measured the percentage of cells containing at least one nuclear focus for each marker (e.g., an Mxc, FLASH, or Mute index). We next determined how many standard deviations these measurements were from the mean index for each marker in control dsRNA treatments (Table S2, z score). The results are summarized in Table I. Although control cells contain robust HLBs as assessed by each marker, all other HLB proteins as well as the MPM-2 epitope fail to form nuclear foci in Mxc-depleted cells (Fig. 6, A–F). Similar results were obtained with FLASH (Table I), suggesting that Mxc and FLASH are critical for HLB assembly.
Figure 6.
HLB formation is disrupted in Mxc-depleted cells. (A–F) S2 cells treated with control (A, C, and E) or mxc (B, D, and F) dsRNAs were stained with MPM-2 (A, B, E, and F), α-Lsm11 (A and B), α-Spt6 (C and D), or α–Mute-LS (E and F) and DAPI. Bars, 2 µm. Note that DAPI staining is not uniform, and under the conditions of imaging the entire nucleus is not always visible. (G–J) Sibling control (G and I) and mxcG48 mutant (H and J) embryos stained with α-Mxc, DAPI, and α–Mute-LS (G and H) or α-FLASH (I and J). Arrows indicate HLBs, and arrowheads indicate an incomplete HLB. Bars, 10 µm. (K) Hierarchical model of Drosophila HLB assembly. Tier 1 and tier 2 are independent of histone gene expression, and HLB localization of tier 2 factors depends on tier 1 factors. Components above the horizontal dotted line are recruited to the HLB in response to particular signals: e.g., transcription for Spt6 and cyclin E/Cdk2 activity for the MPM-2 epitope. Signals that recruit coilin to the HLB are not known. The vertical dotted lines indicate no known interdependency between these factors for HLB localization. The line connecting MPM-2 and Mxc indicates that MPM-2 antibodies bind Mxc.
Our data support the idea that some aspects of HLB assembly are hierarchical. First, knockdown of FLASH and Mxc impaired assembly of Mute, whereas Mute knockdown had no effect on the assembly of FLASH and Mxc. Second, dsRNAs targeting the uncharacterized genes CG9769 and CG7597 reduce detection of Mute and MPM-2 in HLBs but not of Mxc or FLASH. Because Mute, CG9769, and CG7597 are required for the localization and/or MPM-2 reactivity of only a subset of known HLB factors, we suggest that ordered recruitment of individual factors might contribute to HLB assembly (Fig. 6 K).
To extend these observations in vivo, we analyzed HLB assembly in the epidermis of late-stage mxcG48 homozygous mutant embryos. Anti-Mxc staining of HLBs is dramatically reduced in stage 15 mxcG48 embryos but is not absent, likely a result of perdurance of maternal Mxc protein and/or mRNA (Fig. 6, G–J). Nonetheless, we did not detect HLB localization of Mute in mxcG48 embryos (Fig. 6, G and H), and FLASH staining is substantially reduced (Fig. 6, I and J). These data are consistent with Mxc being necessary for proper HLB assembly. Moreover, because all examined HLB markers failed to form nuclear foci in embryos or S2 cells after depletion of Mxc, our data suggest that HLBs do not form in the absence of Mxc.
HLB assembly occurs independently of histone gene expression.
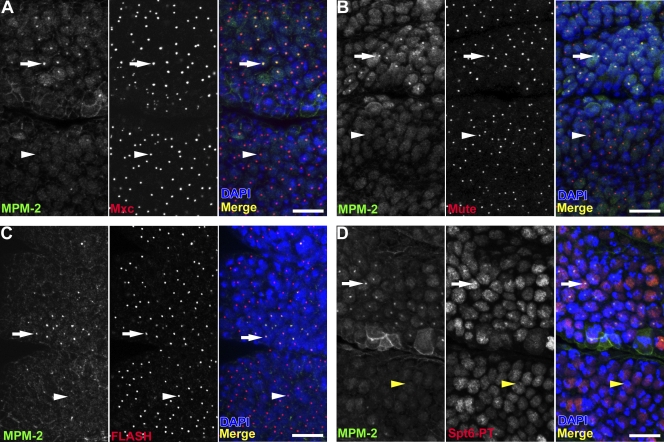
Our biochemical and genetic screening data provided us with two new HLB components (Mxc and Spt6) that together with MPM-2, FLASH, Mute, and U7 snRNP allowed us to further explore the requirements for HLB assembly. We first addressed the relationship between histone gene expression and HLB localization of Mxc, Mute, FLASH, and Spt6. If HLB assembly requires histone gene expression, HLBs should not form in G1-arrested cells, which do not express histone mRNA. To assess this, we took advantage of the known cell cycle program of Drosophila embryogenesis. The first 16 cycles occur with constitutive cyclin E/Cdk2 activity (Sauer et al., 1995) and lack G1 phase, which first appears during the 17th embryonic cycle after cyclin E/Cdk2 is down-regulated (Knoblich et al., 1994). The HLB MPM-2 epitope, which is dependent on cyclin E/Cdk2 activity, is continuously present in interphase nuclei during cycles 14–16 but is absent from epidermal cell nuclei arrested in G1 of cell cycle 17 (Fig. 7, A–D). Like MPM-2, the Mxc, Mute, and FLASH proteins localize to HLBs of all cells in cycles 14–16, but unlike MPM-2, these proteins also localize to HLBs in G1 of cell cycle 17 (Fig. 7, A–C). This result indicates that Mxc, Mute, and FLASH do not require cyclin E/Cdk2 activity or the expression of histone mRNA for localization to HLBs, similar to previous observations showing that the U7 snRNP proteins Lsm10 and Lsm11 form foci in cells that are not in S phase (Liu et al., 2006; White et al., 2007).
Figure 7.
Spt6, but not Mxc, FLASH, or Mute, localization to HLBs is cell cycle dependent. (A–C) Stage 12 w1118 embryos were stained with MPM-2, DAPI, and α-Mxc (A), α–Mute-LS (B), or α-FLASH (C). (D) An embryo similarly stained using α-GFP to detect the Spt6 protein trap fusion protein. Arrows indicate S-phase cells containing MPM-2 foci that colocalize with Mxc (A), Mute-LS (B), FLASH (C), and Spt6-EGFP (D). White arrowheads indicate G117 cells lacking MPM-2 foci but containing Mxc (A), Mute-LS (B), and FLASH (C) in HLBs. Yellow arrowheads indicate a G117 cell lacking foci of MPM-2 and Spt6-EGFP. Thoracic segments 1 and 2 are shown with anterior at the top and ventral on the left. Bars, 10 µm.
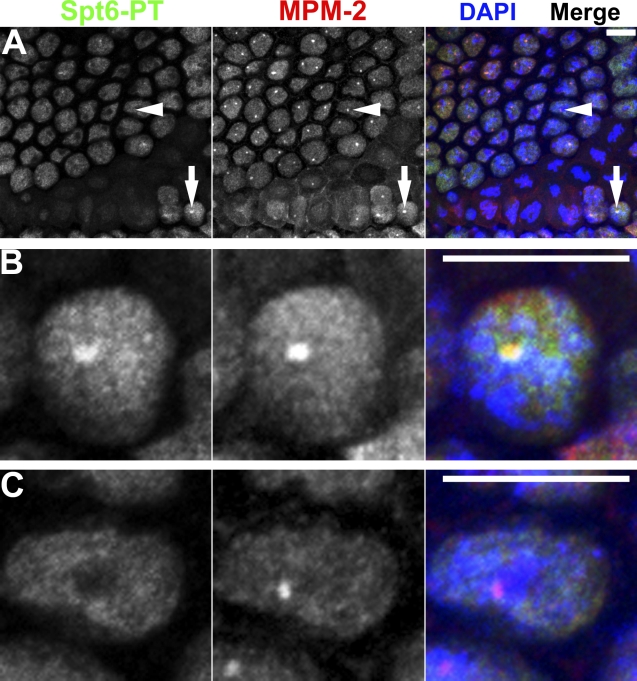
In contrast, using a transgenic line expressing an Spt6-EGFP fusion protein (Buszczak et al., 2007), we find that Spt6 localization to HLBs is dynamic and correlates with active histone gene transcription. First, G1-arrested cells in cycle 17 lack Spt6 foci (Fig. 7 D, arrowheads). Second, we detect prominent Spt6 foci in late G2 cells of cycle 14 at a time when nascent histone transcripts are present in anticipation of the S phase that occurs immediately after completion of mitosis 14 (Fig. 8, A [arrows] and B; Lanzotti et al., 2004). Spt6 foci are not detected in early cycle 14 G2 cells, which do not transcribe histone genes (Fig. 8, A [arrowheads] and C). These data suggest that Spt6 localization to HLBs is coupled to histone gene transcription. Because early G2 cells contain MPM-2 foci (Fig. 8 A, arrows), these data also suggest that cyclin E/Cdk2 activity is not sufficient for Spt6 localization to HLBs. We conclude that some proteins localize to Drosophila HLBs irrespective of histone gene expression, whereas others localize to HLBs only when histone genes are expressed.
Figure 8.
Spt6 localizes to HLBs when histone genes are transcribed. (A) Postblastoderm Spt6-EGFP embryo entering mitosis in cell cycle 14 stained with MPM-2, α-GFP (green), and DAPI. Prominent Spt6 foci form in late G2 cells (arrows) but not in early G2 cells (arrowheads). Single late and early G2 cells are shown in B and C, respectively. Bars, 10 µm.
HLBs remain partially assembled throughout mitosis.
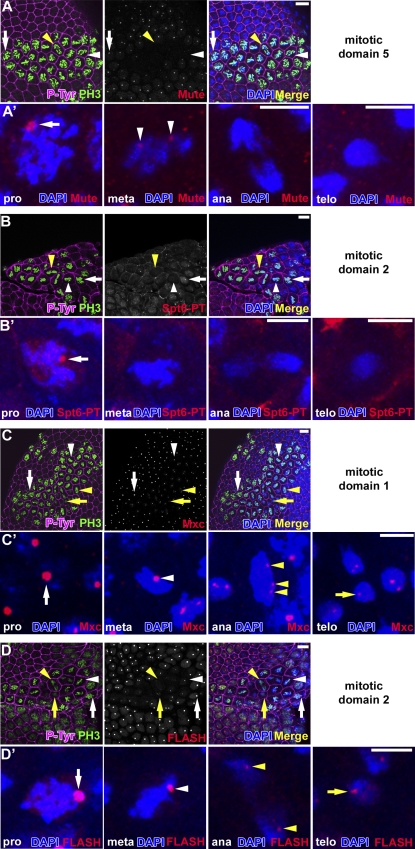
Some nuclear bodies disassemble during mitosis and reform in the subsequent interphase, possibly in response to the resumption of gene expression (Dundr and Misteli, 2010). We previously observed that foci of MPM-2 and Lsm11 disappear at the metaphase–anaphase transition and reappear in the following interphase (White et al., 2007). To determine whether FLASH, Mxc, Mute, and Spt6 undergo a disassembly/assembly cycle, we used antibodies to detect these proteins in domains of mitotic cells of embryonic cell cycle 14 (Fig. 9 and Fig. S2). Similar to Lsm11 and the MPM-2 epitope, Mute foci are present in prophase and some metaphase cells but are not detectable in anaphase cells (Fig. 9 A and Fig. S2 A). Spt6 foci are observed in prophase cells but not in metaphase or anaphase cells (Fig. 9 B and Fig. S2 B). Strikingly, both Mxc and FLASH foci are associated with chromosomes throughout all phases of mitosis (Fig. 9, C and D; Fig. S2, C and D; and Fig. S3). Frequently, we observe three or four small DNA-associated foci of Mxc and FLASH in metaphase and anaphase cells, which is consistent with the detection of individual chromatids. Because Mxc is recognized by MPM-2, the disappearance of MPM-2 foci during mitosis likely represents Mxc dephosphorylation. The close association of Mxc and FLASH with chromosomes during mitosis is not exclusive to early, rapid embryonic cell cycles lacking G1 phase, as Mxc remains associated with chromosomes during mitosis in the canonical cell cycles of postembryonic neuroblasts (Fig. S3, A–D) and in embryonic epidermal mitosis 16, which precedes G1 arrest (Fig. S3, E–H).
Figure 9.
Mxc and FLASH remain chromosome associated during mitosis. (A–D) Postblastoderm w1118 embryos in cell cycle 14 stained with α–P-Tyr, α-PH3 (marks condensed, mitotic chromatin), DAPI, and α-Mute-LS (A), α-GFP (B), α-Mxc (C), or α-FLASH (D). Prophase (pro; white arrows), metaphase (meta; white arrowheads), anaphase (ana; yellow arrowheads), and telophase (telo; yellow arrows) cells are indicated. Bars, 10 µm. (A′–D′) Single prophase, metaphase, anaphase, and telophase cells are shown in A′, B′, C′, and D′. Bars, 5 µm. Note that the mouse α-PH3 used in D stains interphase cells weakly.
Sequential localization of HLB components in the early embryo.
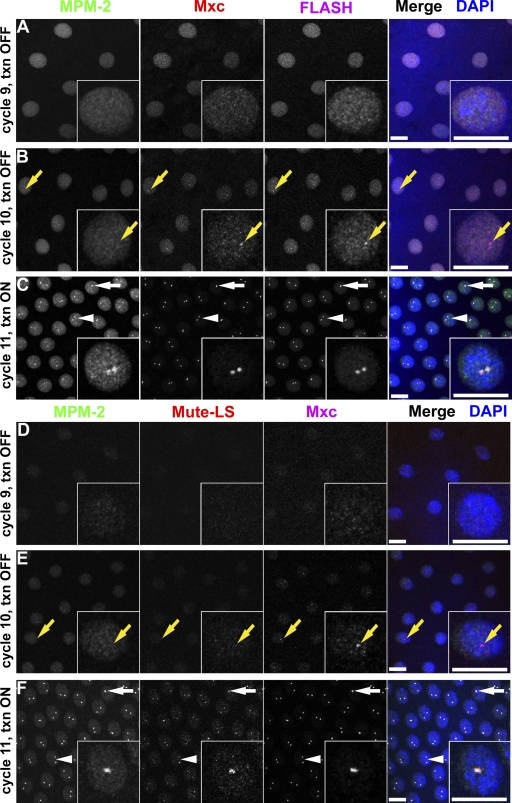
Our observation that Mxc and FLASH are chromosome associated throughout the cell cycle suggests that one or both proteins may directly interact with chromatin at the histone locus. A prediction of this hypothesis is that Mxc and FLASH would be among the first proteins to localize to HLBs during early development. To test this, we examined the timing of appearance of foci of HLB components in the syncytial cycles of early embryos using nuclear density to accurately stage the embryos with respect to each cycle. There are no HLBs in the early syncytial cycles, and MPM-2 and Lsm11 foci are first detected during cycle 11 (Fig. 10), which is precisely when zygotic histone gene expression begins (Edgar and Schubiger, 1986). During nuclear cycle 10, the embryo acquires competence for transcriptional activation of the zygotic genome (Edgar and Schubiger, 1986). Strikingly, we observe colocalizing foci of FLASH and Mxc in cycle 10, one cycle earlier than the appearance of MPM-2/Mute/FLASH/Mxc foci (Fig. 10, B and E), indicating that not all HLB components assemble into nuclear bodies simultaneously during development.
Figure 10.
HLB assembly begins during nuclear cycle 10. (A–C) w1118 syncytial blastoderm embryos stained with MPM-2 (left), α-Mxc (middle), α-FLASH (right), and DAPI. (D–F) w1118 syncytial blastoderm embryos stained with MPM-2 (left), α–Mute-LS (middle), α-Mxc (right), and DAPI. Insets show a single nucleus. Interphase of nuclear cycles 9–11 is shown. Yellow arrows indicate a nucleus containing foci of Mxc and FLASH (B) or Mute (E) lacking a MPM-2 focus. Nuclei with one (white arrows) or two (white arrowheads) HLBs are indicated. txn, histone transcription. Bars, 10 µm.
Discussion
Determining the mechanisms of nuclear body assembly and function is important for understanding how genomes express and maintain genetic information. In this study, we identify a total of 35 factors that when depleted by RNAi either disrupt HLB assembly, histone pre-mRNA processing, or both (Table I). Using several cytological and genetic assays, we conclude that (a) essential components of Drosophila HLBs form bodies independently of histone gene expression, and (b) Drosophila HLBs form by hierarchical recruitment of components.
Drosophila HLBs form independently of histone gene expression
Nuclear bodies are categorized as activity independent or activity dependent (Dundr and Misteli, 2010). In many cases, transcription and processing of RNAs are the associated activities, suggesting that nuclear body formation can be coupled to gene expression. What is striking about Drosophila HLBs is their persistence in cells that are not replicating and, thus, not expressing histone genes. At least four components, Mxc, Mute, FLASH, and U7 snRNP, are present in HLBs in Drosophila embryonic cells that have exited the cell cycle. Liu et al. (2006) reached a similar conclusion for U7 snRNP by examining HLBs in postembryonic cells. Similarly, mammalian HLBs associated with the major histone gene cluster persist in serum-starved cells as judged by NPAT foci (Ye et al., 2003; Ghule et al., 2008).
In contrast, our data show that Spt6 localizes to HLBs only when histone genes are transcribed. Consequently, by this criterion, HLBs would be considered activity dependent. Recruitment to HLBs of Spt6 and possibly other members of the transcription or processing machinery might reflect local accumulation at sites of high transcriptional activity, which occurs at the heat shock locus in response to a heat shock stimulus (Saunders et al., 2006). We show that Spt6 precipitates with MPM-2 in a phosphorylation-dependent manner, suggesting that another mechanism for Spt6 HLB localization might involve a specific interaction with a phosphorylated form of Mxc or another HLB protein.
Consistent with the notion that Drosophila HLBs are not strictly activity dependent, Mxc and FLASH first colocalize into foci during syncytial nuclear cycle 10, one cycle before zygotic histone transcription begins. Their association might be required for subsequent activation of histone gene expression. Mxc and FLASH also persist as colocalizing foci through all stages of mitosis, a time in the cell cycle when transcription is terminated and nascent transcripts are aborted (Shermoen and O’Farrell, 1991). This observation differs from previous studies in mammalian cells that report the disappearance of NPAT foci during mitosis (Ma et al., 2000; Zhao et al., 2000; Ghule et al., 2008).
Multiple mechanisms contribute to Drosophila HLB assembly
The prevailing model of nuclear body biogenesis invokes a process of self-organization of components, but whether such self-organization is entirely stochastic or involves a hierarchical relationship among components is a matter of debate (Misteli, 2001, 2007; Matera et al., 2009; Dundr and Misteli, 2010; Rajendra et al., 2011). Our data suggest that aspects of both mechanisms participate in Drosophila HLB formation. If stochastic self-organization of individual components is sufficient for body formation, the absence of any one component should not affect the assembly of any other component into a body. If hierarchical self-organization plays a role in body formation, the localization of some factors will depend on the presence of others. We previously showed that loss of the variant histone H2aV prevents formation of Lsm11 foci but not of MPM-2 foci (Wagner et al., 2007). Here, we show that depletion of Mxc or FLASH prevents Mute and Lsm11 from forming nuclear foci. These genetic data suggest that stochastic self-organization alone is an incomplete description of Drosophila HLB formation and that hierarchical self-organization plays a role.
In the early embryo, Mxc/FLASH nuclear foci appear during cycle 10, whereas Mxc/FLASH/U7 snRNP/Mute/MPM-2 foci appear in cycle 11. We cannot definitively conclude that the foci in cycle 10 give rise to the foci in cycle 11. Thus, one interpretation of these observations is that two different nuclear bodies are forming, possibly by different mechanisms. However, given the correlation to the timing of the onset of zygotic histone transcription in cycle 11, we consider most parsimonious an interpretation in which the formation of Mxc/FLASH foci represents an early step in HLB assembly that is necessary for subsequent recruitment of other HLB components. The persistence of Mxc/FLASH foci during mitosis while Spt6, Mute, Lsm11, and MPM-2 foci disappear is consistent with this interpretation.
We therefore propose that Drosophila HLB formation involves hierarchical tiers of assembly (Fig. 6 K). Mxc and FLASH form the foundation of this hierarchy, and their assembly into HLB foci is not affected by the removal of other components: Mxc and FLASH colocalize into HLBs after loss of Mute in S2 cells and in Lsm11 mutant flies (Burch et al., 2011). Some observations of mammalian HLBs fit this model. Coilin localization to HLBs is disrupted in the absence of NPAT (Ye et al., 2003). FLASH is required for formation of NPAT foci and vice versa, and in cells in which coilin colocalizes to HLBs with NPAT and FLASH, coilin knockdown does not disrupt HLBs (Barcaroli et al., 2006b). Similarly, Drosophila coilin is present in the HLB of some but not all cells, and coilin mutants do not disrupt HLB formation (Liu et al., 2006, 2009).
By tethering individual components of CBs or HLBs to a specific chromosomal location in mammalian cells, Kaiser et al. (2008) and Shevtsov and Dundr (2011) provide data in support of the stochastic self-organization model of nuclear body formation. These results are not necessarily in conflict with our results. First, the tethering strategy indicates what can happen, not what does happen normally. Perhaps HLB assembly is normally hierarchical and involves some order of assembly, but upon manipulation, the order of assembly can change because the properties of stochastic self-organization of individual components can stimulate body assembly from multiple starting points. MPM-2 and Lsm11 form nuclear foci in Drosophila in the absence of histone genes, and this might reflect self-organizing properties resulting from stochastic interactions (White et al., 2007). FLASH can bind itself (Kiriyama et al., 2009), and we found that the two forms of Mute interact. Thus, we propose a model in which hierarchical aspects of HLB assembly are driven by the set of possible interactions of individual molecules in a given genotype (i.e., wild type vs. mutant/knockdown) and during particular times in development and cell cycle progression. In a recent study, the tethering of paraspeckle protein components to a chromosome did not result in full paraspeckle formation. Rather, active transcription of the Men ε/β noncoding RNA is proposed to “seed” paraspeckle formation (Mao et al., 2011). Similarly, Mxc/FLASH may seed HLB formation.
Mxc: a novel participant in HLB assembly and histone mRNA biosynthesis
Our data suggest that Mxc is the functional equivalent of human NPAT. We find that Mxc likely participates in both histone gene transcription and pre-mRNA processing and could possibly function to coordinate these processes. The essential role we describe for Mxc in HLB assembly supports the idea that HLBs are important for histone mRNA synthesis.
One of the more provocative findings of our study is that the mxc locus encodes a key HLB protein. mxc was named for leg bristle duplications displayed by hypomorphic mutants and is considered a member of the polycomb group of regulators that repress homeotic gene expression during development (Santamaría and Randsholt, 1995; Saget et al., 1998). The ectopic expression of homeotic genes (e.g., Ubx) in mxc mutants (Saget et al., 1998) may result from a failure to synthesize sufficient replication-dependent histones, which are needed to form repressive chromatin (Ner et al., 2002). The cell-autonomous defect in cell proliferation caused by mxc-null mutations is also consistent with a role for Mxc in histone expression (Docquier et al., 1996; Remillieux-Leschelle et al., 2002). Hypomorphic mxc alleles also cause hyperplasia of certain blood cell types that is suppressed by loss of Toll pathway signaling (Remillieux-Leschelle et al., 2002). Such phenotypic pleiotropy suggests that Mxc may regulate genes other than those encoding histones. These issues and the mechanisms of HLB assembly will benefit from further analysis of Drosophila Mxc.
Materials and methods
RNAi screening
In a typical culture of asynchronously proliferating S2 cells, the MPM-2 index is 51% (n = 298 cells). To enhance our ability to detect reduction of the MPM-2 index after dsRNA treatment, we enriched for S phase by incubating cells with aphidicolin and hydroxyurea for 24 h, which increases the MPM-2 index to >90% (n = 53 cells) because cells arrest in S phase with active cyclin E/Cdk2. Under these conditions, a typical cyclin E dsRNA treatment results in an MPM-2 index of 18% (n = 519) compared with 89% (n = 532) for cells treated with control dsRNA. For primary screening, 25 × 104 cells/well were incubated in serum-free media (Sf-900 II SFM; Invitrogen) in 96-well plates containing 1 µg dsRNA/well from a library of 15,680 dsRNAs (Thermo Fisher Scientific) for 6 d at 25°C and treated with 10 µM aphidicolin and 1 µM hydroxyurea during the last 24 h. Cells were transferred for 4–6 h to tissue culture-grade plastic 96-well plates (primary screen) or to Whatman 96-well glass-bottom plates (secondary screens) pretreated with concanavalin A to induce spreading. Cells were fixed in 4% paraformaldehyde for 10 min, extracted using 0.1% Triton X-100 for 15 min, and blocked with 5% normal goat serum for 30 min. Validation/secondary screen dsRNAs were purchased from the Drosophila RNAi Screening Center (Harvard Medical School, Boston, MA). For controls, each plate contained one well with cyclin E dsRNA, two wells with dsRNA from pBluescript (called sk in Fig. 2, A and B), and one well with thread dsRNA (Fig. 2 B, first yellow diamond), which induces apoptosis and was used to assess whether dsRNA-mediated knockdown occurred.
Imaging
Cells were imaged at 25°C in 96-well plates using a high content screening platform (Cellomics Arrayscan VTI [Thermo Fisher Scientific]; 20×/0.4 NA Plan-Neofluar [Carl Zeiss] with a camera; Orca2 ER [Hamamatsu]) and Spotdetector plate protocol (Thermo Fisher Scientific). Confocal images were obtained at 25°C at a zoom of 1.0–8.0 with a 63×, NA of 1.40, Plan Apochromat objective on a laser-scanning confocal microscope (510; Carl Zeiss). Histone pre-mRNA misprocessing reporter images of live cells in serum-free media (SF-900 II) were obtained at 25°C using a UPlanFL N 10×/0.30 Ph1, NA of 0.3, objective on an inverted fluorescence microscope (IX81-ZDC; Olympus) with a camera (ORCA-R2; Hamamatsu) and Volocity acquisition software (PerkinElmer).
Analysis of primary MPM-2 screen data
The Spotdetector plate protocol was optimized to discriminate a nucleus containing MPM-2 foci from a nucleus containing diffuse MPM-2 staining (often observed in wells lacking MPM-2 foci). Cells were defined as objects by DAPI staining, and within each object (i.e., nucleus), the algorithm identified fluorescent spots and quantified the percentage of objects containing at least one fluorescent spot (MPM-2 or spot index). Data were acquired for ≥400 objects for each well when possible, and wells containing fewer than 200 objects were not scored. MPM-2 indices were plotted for each plate and normalized based on the mean MPM-2 index of the plate. A well with an MPM-2 index of more than three standard deviations (i.e., z score of −3.0) below the plate mean was considered a positive. To select candidates for validation, images corresponding to the top scoring 150 dsRNAs were visually confirmed as lacking MPM-2 nuclear foci to eliminate false positives. Genes previously reported to be required for cell cycle progression (Björklund et al., 2006), which when depleted would reduce the MPM-2 index without necessarily encoding an HLB protein, were not selected for further analysis.
Analysis of secondary screen data
For the 95 selected candidates from the primary MPM-2 screen (Table S1), dsRNAs that targeted different mRNA regions than the dsRNAs used in the primary screen were obtained from the Drosophila RNAi screening center. For anti-Mxc, anti-FLASH, and anti-Mute staining, data acquisition and analysis were performed in the same manner as in the primary MPM-2 screen, except that spot indices were adjusted to the mean score of 6–12 control wells lacking dsRNA rather than to the plate mean. Spot index z scores for each dsRNA treatment were calculated as the standard deviation from the control mean score (Table S2). For instance, a z score of −1.5 indicates a mean spot index value <1.5 standard deviations, i.e., below the control mean.
Protein identification by mass spectrometry
Proteins were immunoprecipitated from 10.4 mg of S phase–arrested S2 nuclear extract diluted to 1 mg/ml in NET buffer (50 mM Tris, pH 7.5, 400 mM NaCl, 5 mM EDTA, and 1% NP-40) supplemented with protease and phosphatase inhibitors with 25 µg MPM-2 or anti-HA antibodies precross-linked to 50 µl protein G beads. Immunocomplexes were washed four times, boiled in SDS-PAGE sample buffer, and run on a 4–15% Tris-HCL minigel (Bio-Rad Laboratories). Proteins were stained with Coomassie G-250 (Invitrogen) overnight at 4°C. Proteins of interest were excised from the gel, digested with trypsin, and analyzed by a mass spectrometry analyzer (4800 MALDI TOF/TOF; Applied Biosystems) at the University of North Carolina Michael Hooker Proteomics Center.
Molecular biology
All gene products were expressed in cell culture or as transgenes using Gateway-compatible vectors (Carnegie Institution). Two overlapping PCR products amplified from clone LD32107 and S2 cell cDNA were ligated together to generate a full-length Mxc cDNA. Mxc and CG2247 open reading frames were cloned into pENTR/D-TOPO (Invitrogen), confirmed by sequencing, and recombined with pAVW (Actin5C promoter and N-terminal GFP) and pAWV (Actin5C promoter and C-terminal GFP). pENTR-Mxc was also recombined with pAFW (Actin5C promoter and N-terminal FLAG), pAWF (Actin5C promoter and C-terminal FLAG), and pUGW (ubiquitin promoter and N-terminal GFP). pUGW was a gift from S. Nowotarski and M. Peifer (University of North Carolina, Chapel Hill, NC). Actp-dSpt6FH (Actin5C promoter and C-terminal FLAG/His) was a gift from J. Lis (Cornell University, New York, NY; Andrulis et al., 2002). S2 cells were transfected in 6-well plates using a nucleofector apparatus (Lonza) and Nucleofector V kit (Lonza). Stable cell lines were generated by cotransfection with pCoHygro (Invitrogen) and growing cells in 250 µg/ml hygromycin B (Invitrogen) for >14 d. Northern blotting was performed as previously described (Godfrey et al., 2006).
IPs
Nuclear extracts were made as follows: cells from 100 ml culture were pelleted at 3,000 rpm for 5 min, washed once with PBS, resuspended in hypotonic buffer A (10 mM Hepes, pH 7.9, 1.5 mM MgCL2, 10 mM KCL, and 0.5 mM DTT), and incubated on ice for 30 min. Cells were then lysed with 40 strokes of a dounce homogenizer, and nuclei were pelleted by spinning at 3,000 rpm and resuspended in 100 µl low salt buffer C (20 mM Hepes, pH 7.9, 1.5 mM MgCL2, 0.5 mM DTT, 0.02 M NaCl, and 25% glycerol). An equivalent volume of high salt buffer C (20 mM Hepes, pH 7.9, 1.5 mM MgCL2, 0.5 mM DTT, 1.2 M NaCl, and 25% glycerol) was then added in 10-µl increments, and nuclei were lysed by incubating on a stir plate with a micro stir bar on ice for 30 min. The resulting nuclear extracts were then spun at 13,500 rpm for 5 min to remove nuclear envelopes and chromatin (Sullivan et al., 2009). Antibodies were covalently cross-linked to protein G beads with dimethyl pimelimidate (Rogers et al., 2009). Nuclear extracts were incubated with antibody-coupled beads overnight at 4°C in NET buffer supplemented with 0.5 µg/ml leupeptin, 0.25 µg/ml pepstatin A, 1.5 µg/ml aprotinin, 575 µM phenylmethanesulfonyl fluoride, 1 µM microcystin-LR, and 312.5 nM okadaic acid.
Drosophila strains
mxc alleles and the P[PTT-GA]Spt6CA07692 protein trap line containing an in-frame EGFP insertion between exons 2 and 3 of Spt6 were previously described (Santamaría and Randsholt, 1995; Buszczak et al., 2007). Mxc transgenic flies were created by injecting y1w1118;PBac[y+-attP-3B]VK00033 embryos (BestGene, Inc.) with a ϕC31-compatible vector containing GFP::Mxc expressed from the ubiquitin promoter in pUGW.
Antibody staining
For larval brain and embryos, the following primary antibodies were used: monoclonal mouse MPM-2, (1:1,000; Millipore), monoclonal mouse antiphospho–histone H3 (Ser10; 1:1,000; Millipore), polyclonal rabbit antiphospho–histone H3 (Ser10; 1:1,000; Millipore), polyclonal rabbit antiphosphotyrosine (1:100; Millipore), monoclonal rat antiphosphotyrosine (1:100; R&D Systems), mouse and rabbit anti-GFP (1:2,000; Abcam), affinity-purified polyclonal rabbit anti-FLASH (1:1,000), polyclonal guinea pig anti–Mute-LS (long and short; 1:1,000), affinity-purified rabbit and guinea pig anti-Mxc (1:2,000), and affinity-purified polyclonal guinea pig anti-Spt6 (1:1,000; gift from F. Winston, Harvard Medical School, Boston, MA; Kaplan et al., 2000). For S2 immunostaining, the primary antibodies were monoclonal mouse anti-Ser/Thr-Pro MPM-2 (1:10,000; Millipore), affinity-purified polyclonal rabbit anti-FLASH (1:5,000; Yang et al., 2009), polyclonal guinea pig anti–Mute-LS (1:1,000; gift from S. Bulchand and B. Chia, Temasek Life Sciences Laboratory, National University of Singapore, Singapore), polyclonal guinea pig anti-Mxc (1:5,000), and polyclonal rabbit anti-Hcf (1:2,000; gift from J. Workman, Stowers Institute for Medical Research, Kansas City, MO; Guelman et al., 2006). The secondary antibodies used (1:1,000) in all experiments were goat anti–mouse IgG labeled with Oregon green 488 (Invitrogen), Cy3 (Jackson ImmunoResearch Laboratories, Inc.), or Cy5 (Jackson ImmunoResearch Laboratories, Inc.); goat anti–rabbit IgG labeled with Rhodamine red (Invitrogen) or Cy2 (Jackson ImmunoResearch Laboratories, Inc.); goat anti–rat IgG labeled with Cy3 and donkey anti–rat Cy5; and goat anti–guinea pig IgG labeled with Alexa Fluor 488 (Invitrogen), Cy3, or Cy5. DNA was detected by staining embryos with DAPI (1:1,000 of 1 mg/ml stock; Dako) for 2 min. Embryos were dechorionated, fixed in a 1:1 mixture of 5% formaldehyde/heptane for 20 min, and incubated with primary and secondary antibodies each for 1 h at 25°C or overnight at 4°C. Brains were dissected from third instar larvae in Schneider’s media, fixed in 4% formaldehyde for 20 min, and permeabilized in 1% Triton X-100 for 1 h before immunostaining.
Reporter assay for histone pre-mRNA misprocessing
Dmel-2 cells stably expressing the Act5Cp/H3/GFP reporter (Yang et al., 2009) were cultured for 5 d in a 96-well plate containing dsRNAs and then imaged. GFP fluorescence/cell from two replicate experiments was quantified from epifluorescence and bright-field images using ImageJ (National Institutes of Health). RGB images were first converted to grayscale. Image brightness was standardized to a common background level for each image to account for variations in field brightness introduced during image acquisition. Gridlines were superimposed over the images to define regions to be scored, three to five representative scoring boxes were selected, and the fluorescence within each of these boxes was calculated by summing the number of black pixels per box. Next, a phase-contrast image of the cells was overlaid with the same grid and individual cells were counted for each box, allowing for a calculation of the amount of fluorescence per cell. These values were normalized to the mean fluorescence per cell of a known positive control (e.g., FLASH) to allow comparison of data from different experiments. Finally, the relative fluorescence per cell derived from two independent experiments was averaged and expressed as a percentage of the value obtained with the positive control.
Online supplemental material
Fig. S1 shows the histone misprocessing reporters used in this study. Fig. S2 shows that Mxc and FLASH remain chromosome associated during mitosis. Fig. S3 shows that Mxc remains chromosome associated during mitosis in embryonic and larval cell division cycles containing G1. Table S1 shows the RNAi screening data used in this study. Table S2 shows the validation and secondary screen z scores. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201012077/DC1.
Acknowledgments
We are grateful to Sarada Bulchand and Bill Chia for generously sharing anti-Mute antibodies and information before publication and Nasser Rusan and Greg Rogers for their help with the RNAi screen. We thank Severine Landais, Leanne Jones, T.K. Rajendra, and Greg Matera for sharing unpublished information, Sarah Anderson for help sequencing mxc alleles, John Lis, Fred Winston, Steph Nowotarski, and Jerry Workman for reagents, Harmony Salzler for discussions about HLB assembly models, and Wade Harper, Steve Rogers, and Mark Peifer for comments on the manuscript.
This work was supported by a Canadian Institute of Health Research grant MDR-85476 to A.E. White and National Institutes of Health grants GM58921 to W.F. Marzluff and Z. Dominski and GM57859 to R.J. Duronio.
Footnotes
Abbreviations used in this paper:
- CB
- Cajal body
- dsRNA
- double-stranded RNA
- FLASH
- FLICE-associated huge protein
- HLB
- histone locus body
- IP
- immunoprecipitation
- LisH
- lissencephaly homology
- Mxc
- multisex combs
- NPAT
- nuclear protein of the ataxia telangiectasia–mutated locus
- SLBP
- stem loop–binding protein
- snRNP
- small nuclear RNP
- Spt6
- suppressor of Ty 6
References
- Andrulis E.D., Guzmán E., Döring P., Werner J., Lis J.T. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635–2649 10.1101/gad.844200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Werner J., Nazarian A., Erdjument-Bromage H., Tempst P., Lis J.T. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 420:837–841 10.1038/nature01181 [DOI] [PubMed] [Google Scholar]
- Ardehali M.B., Yao J., Adelman K., Fuda N.J., Petesch S.J., Webb W.W., Lis J.T. 2009. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 28:1067–1077 10.1038/emboj.2009.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaroli D., Bongiorno-Borbone L., Terrinoni A., Hofmann T.G., Rossi M., Knight R.A., Matera A.G., Melino G., De Laurenzi V. 2006a. FLASH is required for histone transcription and S-phase progression. Proc. Natl. Acad. Sci. USA. 103:14808–14812 10.1073/pnas.0604227103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaroli D., Dinsdale D., Neale M.H., Bongiorno-Borbone L., Ranalli M., Munarriz E., Sayan A.E., McWilliam J.M., Smith T.M., Fava E., et al. 2006b. FLASH is an essential component of Cajal bodies. Proc. Natl. Acad. Sci. USA. 103:14802–14807 10.1073/pnas.0604225103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björklund M., Taipale M., Varjosalo M., Saharinen J., Lahdenperä J., Taipale J. 2006. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 439:1009–1013 10.1038/nature04469 [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L., De Cola A., Vernole P., Finos L., Barcaroli D., Knight R.A., Melino G., De Laurenzi V. 2008. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 7:2357–2367 [DOI] [PubMed] [Google Scholar]
- Bulchand S., Menon S.D., George S.E., Chia W. 2010. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J. Cell Sci. 123:2697–2707 10.1242/jcs.063172 [DOI] [PubMed] [Google Scholar]
- Burch B.D., Godfrey A.C., Gasdaska P.Y., Salzler H.R., Duronio R.J., Marzluff W., Dominski Z. 2011. The interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A.D., Nystul T.G., Ohlstein B., Allen A., et al. 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics. 175:1505–1531 10.1534/genetics.106.065961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Jin J., Swanson S.K., Cole M.D., Choi S.H., Florens L., Washburn M.P., Conaway J.W., Conaway R.C. 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 285:4268–4272 10.1074/jbc.C109.087981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B.R., Lilly M.A., Spradling A.C. 1998. Cell cycle control of chorion gene amplification. Genes Dev. 12:734–744 10.1101/gad.12.5.734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis F.M., Tsao T.Y., Fowler S.K., Rao P.N. 1983. Monoclonal antibodies to mitotic cells. Proc. Natl. Acad. Sci. USA. 80:2926–2930 10.1073/pnas.80.10.2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRan M., Pulvino M., Greene E., Su C., Zhao J. 2008. Transcriptional activation of histone genes requires NPAT-dependent recruitment of TRRAP-Tip60 complex to histone promoters during the G1/S phase transition. Mol. Cell. Biol. 28:435–447 10.1128/MCB.00607-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docquier F., Saget O., Forquignon F., Randsholt N.B., Santamaria P. 1996. The multi sex combs gene of Drosophila melanogaster is required for proliferation of the germline. Roux’s Arch. Dev. Biol. 205:203–214 10.1007/BF00365798 [DOI] [PubMed] [Google Scholar]
- Dominski Z., Marzluff W.F. 2007. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 396:373–390 10.1016/j.gene.2007.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M., Misteli T. 2010. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2:a000711 10.1101/cshperspect.a000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Schubiger G. 1986. Parameters controlling transcriptional activation during early Drosophila development. Cell. 44:871–877 10.1016/0092-8674(86)90009-7 [DOI] [PubMed] [Google Scholar]
- Frey M.R., Matera A.G. 1995. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. USA. 92:5915–5919 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule P.N., Dominski Z., Yang X.C., Marzluff W.F., Becker K.A., Harper J.W., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S. 2008. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 105:16964–16969 10.1073/pnas.0809273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey A.C., Kupsco J.M., Burch B.D., Zimmerman R.M., Dominski Z., Marzluff W.F., Duronio R.J. 2006. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA. 12:396–409 10.1261/rna.2270406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelman S., Suganuma T., Florens L., Swanson S.K., Kiesecker C.L., Kusch T., Anderson S., Yates J.R., III, Washburn M.P., Abmayr S.M., Workman J.L. 2006. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol. Cell. Biol. 26:871–882 10.1128/MCB.26.3.871-882.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger K.E., Gall J.G. 2006. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 16:19–26 10.1016/j.tcb.2005.11.005 [DOI] [PubMed] [Google Scholar]
- Huang A.M., Rubin G.M. 2000. A misexpression screen identifies genes that can modulate RAS1 pathway signaling in Drosophila melanogaster. Genetics. 156:1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T.E., Intine R.V., Dundr M. 2008. De novo formation of a subnuclear body. Science. 322:1713–1717 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- Kaplan C.D., Morris J.R., Wu C., Winston F. 2000. Spt5 and spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623–2634 10.1101/gad.831900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriyama M., Kobayashi Y., Saito M., Ishikawa F., Yonehara S. 2009. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol. Cell. Biol. 29:4729–4741 10.1128/MCB.00289-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A., Sauer K., Jones L., Richardson H., Saint R., Lehner C.F. 1994. Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell. 77:107–120 10.1016/0092-8674(94)90239-9 [DOI] [PubMed] [Google Scholar]
- Lanzotti D.J., Kupsco J.M., Marzluff W.F., Duronio R.J. 2004. string(cdc25) and cyclin E are required for patterned histone expression at different stages of Drosophila embryonic development. Dev. Biol. 274:82–93 10.1016/j.ydbio.2004.06.019 [DOI] [PubMed] [Google Scholar]
- Liu J.L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J.G. 2006. The Drosophila melanogaster Cajal body. J. Cell Biol. 172:875–884 10.1083/jcb.200511038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.L., Wu Z., Nizami Z., Deryusheva S., Rajendra T.K., Beumer K.J., Gao H., Matera A.G., Carroll D., Gall J.G. 2009. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol. Biol. Cell. 20:1661–1670 10.1091/mbc.E08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T., Van Tine B.A., Wei Y., Garrett M.D., Nelson D., Adams P.D., Wang J., Qin J., Chow L.T., Harper J.W. 2000. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 14:2298–2313 10.1101/gad.829500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y.S., Sunwoo H., Zhang B., Spector D.L. 2011. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13:95–101 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff W.F., Wagner E.J., Duronio R.J. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9:843–854 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., Izaguire-Sierra M., Praveen K., Rajendra T.K. 2009. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev. Cell. 17:639–647 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., Taipale M., Kind J., Holz H., Gebhardt P., Schelder M., Vermeulen M., Buscaino A., Duncan K., Mueller J., et al. 2006. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol. Cell. 21:811–823 10.1016/j.molcel.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Miele A., Braastad C.D., Holmes W.F., Mitra P., Medina R., Xie R., Zaidi S.K., Ye X., Wei Y., Harper J.W., et al. 2005. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol. Cell. Biol. 25:6140–6153 10.1128/MCB.25.14.6140-6153.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2001. The concept of self-organization in cellular architecture. J. Cell Biol. 155:181–185 10.1083/jcb.200108110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. 2007. Beyond the sequence: cellular organization of genome function. Cell. 128:787–800 10.1016/j.cell.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Ner S.S., Harrington M.J., Grigliatti T.A. 2002. A role for the Drosophila SU(VAR)3-9 protein in chromatin organization at the histone gene cluster and in suppression of position-effect variegation. Genetics. 162:1763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Will C.L., Lührmann R., Schümperli D., Müller B. 2001. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 20:5470–5479 10.1093/emboj/20.19.5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai R.S., Grimmler M., Meister G., Will C.L., Lührmann R., Fischer U., Schümperli D. 2003. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17:2321–2333 10.1101/gad.274403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel M., Feller C., Straub T., Mitlöhner H., Becker P.B. 2010. The activation potential of MOF is constrained for dosage compensation. Mol. Cell. 38:815–826 10.1016/j.molcel.2010.05.022 [DOI] [PubMed] [Google Scholar]
- Rajendra T.K., Praveen K., Matera A.G. 2011. Genetic analysis of nuclear bodies: from nondeterministic chaos to deterministic order. Cold Spring Harb. Symp. Quant. Biol. 10.1101/sqb.2010.75.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remillieux-Leschelle N., Santamaria P., Randsholt N.B. 2002. Regulation of larval hematopoiesis in Drosophila melanogaster: a role for the multi sex combs gene. Genetics. 162:1259–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Roberts D.M., Peifer M., Rogers S.L. 2009. The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saget O., Forquignon F., Santamaria P., Randsholt N.B. 1998. Needs and targets for the multi sex combs gene product in Drosophila melanogaster. Genetics. 149:1823–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría P., Randsholt N.B. 1995. Characterization of a region of the X chromosome of Drosophila including multi sex combs (mxc), a Polycomb group gene which also functions as a tumour suppressor. Mol. Gen. Genet. 246:282–290 10.1007/BF00288600 [DOI] [PubMed] [Google Scholar]
- Sauer K., Knoblich J.A., Richardson H., Lehner C.F. 1995. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes Dev. 9:1327–1339 10.1101/gad.9.11.1327 [DOI] [PubMed] [Google Scholar]
- Saunders A., Core L.J., Lis J.T. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7:557–567 10.1038/nrm1981 [DOI] [PubMed] [Google Scholar]
- Shermoen A.W., O’Farrell P.H. 1991. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 67:303–310 10.1016/0092-8674(91)90182-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov S.P., Dundr M. 2011. Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13:167–173 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- Shibutani S.T., de la Cruz A.F., Tran V., Turbyfill W.J., III, Reis T., Edgar B.A., Duronio R.J. 2008. Intrinsic negative cell cycle regulation provided by PIP box- and Cul4Cdt2-mediated destruction of E2f1 during S phase. Dev. Cell. 15:890–900 10.1016/j.devcel.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T., Gutiérrez J.L., Li B., Florens L., Swanson S.K., Washburn M.P., Abmayr S.M., Workman J.L. 2008. ATAC is a double histone acetyltransferase complex that stimulates nucleosome sliding. Nat. Struct. Mol. Biol. 15:364–372 10.1038/nsmb.1397 [DOI] [PubMed] [Google Scholar]
- Sullivan E., Santiago C., Parker E.D., Dominski Z., Yang X., Lanzotti D.J., Ingledue T.C., Marzluff W.F., Duronio R.J. 2001. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev. 15:173–187 10.1101/gad.862801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K.D., Steiniger M., Marzluff W.F. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell. 34:322–332 10.1016/j.molcel.2009.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Burch B.D., Godfrey A.C., Salzler H.R., Duronio R.J., Marzluff W.F. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell. 28:692–699 10.1016/j.molcel.2007.10.009 [DOI] [PubMed] [Google Scholar]
- Wei Y., Jin J., Harper J.W. 2003. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol. Cell. Biol. 23:3669–3680 10.1128/MCB.23.10.3669-3680.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White A.E., Leslie M.E., Calvi B.R., Marzluff W.F., Duronio R.J. 2007. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol. Biol. Cell. 18:2491–2502 10.1091/mbc.E06-11-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.H., Gall J.G. 1993. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc. Natl. Acad. Sci. USA. 90:6257–6259 10.1073/pnas.90.13.6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.C., Burch B.D., Yan Y., Marzluff W.F., Dominski Z. 2009. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol. Cell. 36:267–278 10.1016/j.molcel.2009.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Wei Y., Nalepa G., Harper J.W. 2003. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol. Cell. Biol. 23:8586–8600 10.1128/MCB.23.23.8586-8600.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Dynlacht B., Imai T., Hori T., Harlow E. 1998. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes Dev. 12:456–461 10.1101/gad.12.4.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Kennedy B.K., Lawrence B.D., Barbie D.A., Matera A.G., Fletcher J.A., Harlow E. 2000. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 14:2283–2297 10.1101/gad.827700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Roeder R.G., Luo Y. 2003. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell. 114:255–266 10.1016/S0092-8674(03)00552-X [DOI] [PubMed] [Google Scholar]