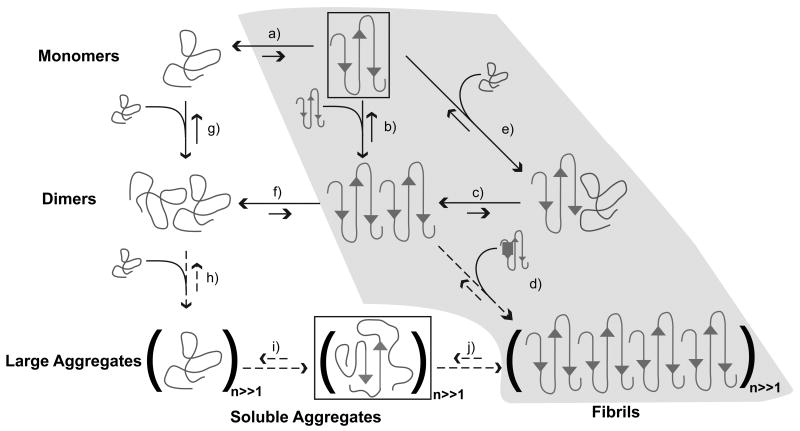
Figure 4. Schematic of possible aggregation pathways for polyglutamine in vitro.
Here, n denotes the number of polyglutamine molecules within a disordered aggregate and nF denotes the number of polyglutamine molecules within an ordered amyloid fibril. The ordered amyloid fibril rich in β-sheets is shown in the bottom right corner of the schematic. The gray shaded region encompasses steps (a), (e), (c), and (d) and depicts the homogeneous nucleation proposal of Chen et al. [83]. Studies of the thermodynamics of step (a), indicate that the formation of ordered conformations is thermodynamically unfavorable. Monomeric polyglutamine prefers disordered, collapsed conformations, left of step (a). Step (b) pertains to the thermodynamics of interactions between chains that adopt ordered conformations. However, the likelihood that chains will sample the associations shown in step (b) is very small because this is tied to the equilibria in step (a), which requires the population of the conformations with high β-content. Similarly, step (f) shows that disordered dimers are thermodynamically favored to dimers with high β-contents in individual chains. This is the result of linkage to step (a) as discussed above. The aggregates achieved in step (h) are likely to be large (in terms of n) and exhibit spherical, “liquid-like” [102-104] organization of polyglutamine chains around each other. Step (i) depicts a slow conformational conversion of individual / small numbers of chains to β-sheets. This slow step is likely to lead to the creation of an ordered template for fibril formation via monomer or oligomer addition and elongation to yield the ordered amyloid fibril.