Table 1.
The RecQ helicase can bind to and unwind a wide variety of DNA substrates
| DNA substrate
|
Structure (annealed oligonucleotides)a
|
Unwinding rateb (nm (bp)/nm RecQ/sec)
|
Relative binding affinityc
|
|---|---|---|---|
| 3-way junction | 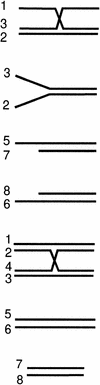 |
2.9 | 7 |
| Flayed duplex | 2.2 | 8 | |
| 5′-ssDNA overhang | 1.2 | 2 | |
| 3′-ssDNA overhang | 1.1 | 12 | |
| 4-way junction | 1.0 | 1 | |
| 63-bp blunt duplex | 0.8 | N/D | |
| 48-bp blunt duplex | 0.2 | 1 |
Reactions were performed and analyzed as described in Materials and Methods.
Numbers correspond to the oligonucleotides in Table 2 annealed as described in Materials and Methods to form the indicated DNA substrate.
Calculated from the linear slope of a plot of unwinding rate [nm (bp)/sec] vs. RecQ helicase concentration (nm). The relative error for each rate is approximately ±5%.
Determined as in Materials and Methods and normalized to the binding value of 5 × 106/m obtained for the 48-bp blunt duplex substrate. The relative error for each value is approximately ±5%.
