Abstract
Rationale
Mammalian cardiomyocytes withdraw from the cell cycle during early post-natal development, which significantly limits the capacity of the adult mammalian heart to regenerate following injury. The regulatory mechanisms which govern cardiomyocyte cell cycle withdrawal and binucleation are poorly understood.
Objective
Given the potential of microRNAs (miRNAs) to influence large gene networks and modify complex developmental and disease phenotypes, we searched for miRNAs that were regulated during the postnatal switch to terminal differentiation.
Methods and Results
Microarray analysis revealed subsets of miRNAs that were up- or down-regulated in cardiac ventricles from mice at 1- and 10-days of age (P1 and P10). Interestingly, miR-195 (a member of the miR-15 family) was the most highly up-regulated miRNA during this period, with expression levels almost 6-fold higher in P10 ventricles relative to P1. Precocious over-expression of miR-195 in the embryonic heart was associated with ventricular hypoplasia and ventricular septal defects in βMHC-miR-195 transgenic mice. Using global gene profiling and Argonaute-2 immunoprecipitation approaches, we show that miR-195 regulates the expression of a number of cell cycle genes, including checkpoint kinase 1 (Chek1), which we identify as a highly conserved direct target of miR-195. Finally, we demonstrate that knock-down of the miR-15 family in neonatal mice using locked nucleic acid (LNA)-modified antimiRs is associated with an increased number of mitotic cardiomyocytes and de-repression of Chek1.
Conclusions
These findings suggest that up-regulation of the miR-15 family during the neonatal period may be an important regulatory mechanism governing cardiomyocyte cell cycle withdrawal and binucleation.
Keywords: miR-195, neonate, cell cycle, binucleation, Chek1
Introduction
The adult mammalian heart is one of the least regenerative organs in the body. In contrast, the neonatal mammalian heart possesses a remarkable capacity for cardiac regeneration.1 Importantly, the neonatal mouse heart’s ability to regenerate following injury is lost by 7 days-of-age, a time point coinciding with the onset of cardiomyocyte proliferative arrest and binucleation in rodents.2, 3 Genetic fate-mapping experiments in the neonatal mouse1 and adult zebrafish4, 5 suggest that the majority of regenerated cardiomyocytes are derived from pre-existing cardiomyocytes through cell division, rather than activation of an undifferentiated stem or progenitor cell. Therefore, understanding the mechanisms regulating the mammalian heart’s post-natal transition towards cell cycle arrest is of fundamental importance.
Cardiomyocytes undergo extensive proliferation during embryogenesis, but withdraw from the proliferative cell cycle shortly after birth.6 Beginning at post-natal day 5 in rodents, the majority of cardiomyocytes undergo a final round of DNA replication and karyokinesis in the absence of cytokinesis, which results in binucleation of approximately 90% of cardiomyocytes by the second week of post-natal life.2, 3 Although recent studies suggest that adult cardiomyocytes can re-enter the cell cycle and proliferate in response to certain mitogens (e.g. fibroblast growth factor, periostin, neuregulin),7–9 it appears that this proliferative potential is restricted to a small proportion of mononucleated cardiomyocytes.7 While post-natal activation of tumor suppressor pathways appears to be an important mechanism for cardiomyocyte terminal differentiation,10 the molecular mechanisms regulating post-natal cardiomyocyte binucleation and mitotic arrest remain one of the most poorly understood aspects of cardiac biology.
Recently, microRNAs (miRNAs) have emerged as important regulators of almost every aspect of cardiac biology,11 including cardiomyocyte proliferation.12, 13 MiRNAs are small (~22 nucleotides), non-coding RNAs that induce post-transcriptional repression of gene expression by binding to target recognition sequences predominantly located within the 3’ untranslated region (3’UTR) of target mRNAs.11 Targeting of a miRNA to its mRNA target is dependent on the “seed” sequence, spanning bases 2–8 of the 5’ portion of a mature miRNA. Individual miRNAs can target numerous mRNAs and it is becoming increasingly evident that miRNAs often exert their actions by modestly repressing the expression of many mRNAs with related biological functions.11 Furthermore, some of the most abundant miRNAs in the heart belong to large miRNA ”seed” families, comprising multiple miRNAs that share a high degree of sequence homology and a common “seed” region.14 Therefore, multiple miRNAs with related functions within a miRNA family have the potential to redundantly regulate complex biological processes by targeting multiple genes in a common pathway. Despite the vast regulatory potential of miRNAs and their involvement in various aspects of cell cycle control, little is known about how miRNAs might contribute to post-natal maturation and terminal differentiation of cardiomyocytes.
In an attempt to identify miRNAs involved in cardiomyocyte mitotic arrest, we examined the developmental expression pattern of miRNAs in the heart during the post-natal transition towards binucleation and cell cycle withdrawal. Here, we describe a collection of miRNAs that are dysregulated in the heart between post-natal days 1 and 10. Interestingly, miR-195, a member of the miR-15 family, was the most highly up-regulated miRNA during this period. We show that premature over-expression of miR-195 in the embryonic heart causes a spectrum of congenital heart abnormalities associated with premature cell cycle arrest. miR-195 negatively regulates the expression of a number of cell cycle genes, including checkpoint kinase 1 (Chek1), which was found to be enriched within the RNA-induced silencing complex (RISC) of neonatal miR-195 transgenic hearts in vivo. Post-natal inhibition of the miR-15 family with antimiRs was associated with an increased number of mitotic cardiomyocytes and de-repression of several miR-15 family target genes, including Chek1. These results reveal that up-regulation of the miR-15 family provides a previously unappreciated layer of regulatory control that contributes to post-natal cardiomyocyte cell cycle withdrawal after birth.
Methods
An expanded Methods section is available in the Online Data Supplement at http://circres.ahajournals.org.
Experimental Animals and Tissue Collection
All protocols were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center. C57BL/6 mice (The Jackson Laboratory) were used for miRNA expression profiling during post-natal development. For RNA analysis, atrial tissues were removed from ventricles (septum intact) and ventricles were blotted and weighed before snap-freezing in liquid nitrogen for storage at −80°C.
Generation of βMHC-miR-195 Transgenic Mice
A mouse genomic fragment flanking miR-195 was subcloned into a cardiac-specific expression plasmid containing 7 kb of upstream regulatory sequence from the βMHC promoter and human growth hormone poly(A)+ signal.15 Transgenic mice were generated by pronuclear injection using standard procedures and maintained on a mixed genetic background.
In Vivo Delivery of Synthetic LNA-antimiR Oligonucleotides to Neonatal Mice
Locked nucleic acid (LNA)-modified antimiRs directed against the mature miR-15b and miR-16 sequences were synthesized as unconjugated and fully phosphorothiolated oligonucleotides (miRagen Therapeutics, CO). The LNA-antimiRs were designed to perfectly complement 16 nucleotides of the mature miR-15b and miR-16 sequences, respectively (sequences shown in Supplemental Table 2). A control LNA-antimiR was also used, which contained the same LNA/DNA chemical composition as the anti-15/16 oligos, but was designed to target a miR sequence that is unique to C. elegans and is not expressed in mammals. Neonatal mice (CD-1/ICR strain, Charles River Laboratories) received control oligo, anti-miR-15b, anti-miR-16 or a combination of anti-miR-15b/16 at a dose of 25 mg/kg body weight via subcutaneous injection once per day at post-natal days 2, 3 and 4 (P2–P4). Hearts were harvested at P12 (i.e. 10 days after the first treatment) for assessment of miRNA knockdown, target gene regulation and histology.
Histological Analysis
Hearts used for histology were briefly rinsed in PBS, fixed in 4% paraformaldehyde overnight, and then transferred to 50% ethanol until paraffin embedding. Sections (5 μm thickness) were processed for H&E and Masson’s trichrome staining according to standard procedures.
Immunofluorescence
All immunofluorescence was performed on paraffin sections as previously described.1 Antibody details are in the Online Methods section.
RNA analysis
Total RNA was extracted from mouse hearts using TRIzol (Invitrogen), according to the manufacturer’s instructions Details of real-time PCR analysis and Northern blotting are provided in the Online Methods supplement. Real-time PCR primers and Northern probes are provided in Supplemental Tables 3 and 4, respectively.
Microarray for miRNAs
miRNA profiling of P1 vs. P10 cardiac ventricles from C57BL/6 mice (n=3 per group) was performed using LC Science’s miRNA microarray service (Chip ID = HM13.0). Details of sample preparation and analysis are as previously described.16
Microarray for mRNAs
Microarray analysis was performed on RNA samples extracted from P1 cardiac ventricles from αMHC-miR-195 transgenic and wild-type mice.17 Microarray analysis was performed by the University of Texas Southwestern Microarray Core Facility using the Mouse Genome 430 2.0 Array (Affymetrix, CA) as described.18 A fold change cut-off of 1.5 was considered significant.
RISC RNA Sequencing
RISC RNA sequencing experiments were conducted on Argonaute 2 (Ago2) immunoprecipitates from P1 cardiac ventricles from αMHC-miR-195 transgenic and wild-type mice (n=3 WT and n=4 TG), as described previously.19 A fold enrichment cut-off of greater than 2.0 was considered significant. MiR-15 family binding sites were identified in putative target transcripts by cross-referencing to three different computational algorithms for prediction of miRNA 3’UTR binding sites (TargetScan 5.1, miRanda 3.0 and PITA v6.0).
Gene Ontology Cluster Analysis
Gene ontology analysis was performed for genes that were significantly regulated in miR-195 transgenic hearts (>1.5-fold) and for genes that were enriched in miR-195 transgenic RISC (>2-fold). Gene ontology analysis was conducted using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional annotation tool. DAVID analysis was performed with ‘High’ classification stringency and P-value cut-off of 0.001.
Cell Culture, Transfection and Luciferase Assays
A 1948-bp genomic fragment of the Chek1 3’ UTR encompassing the miR-15 family binding site was PCR-amplified and ligated into the firefly luciferase (f-luc) reporter construct (pMIR-REPORTTM; Ambion). The seed region of the miR-195 target site in the Chek1 3’UTR was mutated using the QuickChange II site-directed mutagenesis kit (Stratagene). Primer sequences are provided in Supplemental Table 5.
Results
Identification of miRNAs Regulated During Post-natal Heart Development
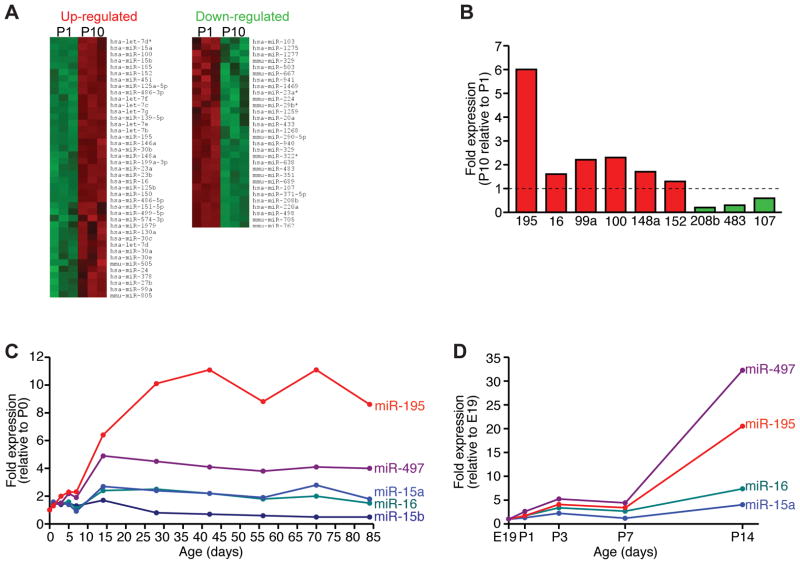
To identify miRNAs involved in the post-natal onset of cardiomyocyte binucleation and cell cycle withdrawal, we compared miRNA expression profiles in mouse cardiac ventricles at post-natal days 1 and 10 (P1 and P10). A total of 71 miRNAs were differentially regulated during this developmental transition (41 miRNAs showed increased expression between P1 and P10 and 30 miRNAs showed decreased expression between P1 and P10) (Figure 1A and Online Table I). One of the most robustly down-regulated miRNAs between P1 and P10 was miR-208b, located within an intron of the Myh7 gene,20 which is well-known to be down-regulated after birth.21 Interestingly, multiple large miRNA families were up-regulated between P1 and P10, including the miR-15, miR-30 and let-7 families (Figure 1A and Online Table I). A subset of miRNAs from the array data were confirmed by real-time PCR analysis using probes specific for the mature miRNA sequences in the mouse (Figure 1B).
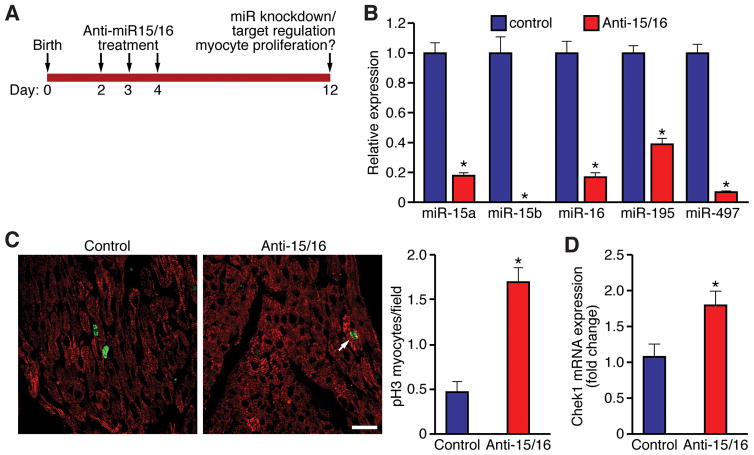
Figure 1. MicroRNA expression profiling during post-natal cardiac development.
A, Heat map showing microRNAs that are either up- or down-regulated in the heart between post-natal days 1 and 10 (P1 and P10) in the mouse. B, Real-time PCR analysis for a subset of microRNAs that are regulated during post-natal cardiac development. miR expression levels are normalized to U6 and represented as a fold change relative to P1 ventricles. C, Temporal regulation of miR-15 family members in the heart during post-natal cardiac development. Expression levels of each miR-15 family member are normalized to U6 and represented as a fold change relative to P0 ventricles. D, Temporal regulation of miR-15 family members in isolated cardiomyocytes. Expression levels of each miR-15 family member are normalized to U6 and represented as a fold change relative to E19 cardiomyocytes.
Multiple miR-15 Family Members are Up-regulated During Post-natal Heart Development
The most highly up-regulated miRNA between P1 and P10 was miR-195 (Figure 1B), a member of the miR-15 family. The miR-15 family consists of 6 highly conserved miRNAs, which are clustered on three separate chromosomes (Online Figure IA). All members of the miR-15 family (miR-15a, miR-15b. miR-16-1, miR-16-2, miR-195 and miR-497) share a common seed region, with varying degrees of sequence homology in the non-seed region of the mature miRNA (Online Figure IB). Northern blot analysis of multiple adult mouse tissues demonstrated miR15a, miR-16, miR-195 and miR-497 expression in most adult tissues, including the heart (Online Figure IIA).
The precise time course of miR-15 family up-regulation was further examined in mouse hearts harvested at multiple time points between P0 and P84. Real-time PCR analysis demonstrated that miR-195, miR-497, miR-15a and miR-16 were all up-regulated in mouse cardiac ventricles between P7 and P14 (Figure 1C), a time point coinciding with the onset of cardiomyocyte binucleation.2, 3 Up-regulation of miR-195, miR-497 and miR-16 between P7 and P14 was confirmed by Northern blot (Online Figure IIB). In order to examine whether the post-natal up-regulation of miR-15 family members occurred in the myocyte population or another cell type, we isolated embryonic and neonatal rat cardiomyocytes between E19 and P14 and profiled miR-15 family expression. Real-time PCR analysis confirmed that multiple miR-15 family members were up-regulated in rodent ventricular cardiomyocytes between P7 and P14 (Figure 1D).
Over-expression of miR-195 in the Developing Heart Causes Congenital Heart Abnormalities Associated with Premature Cell Cycle Arrest
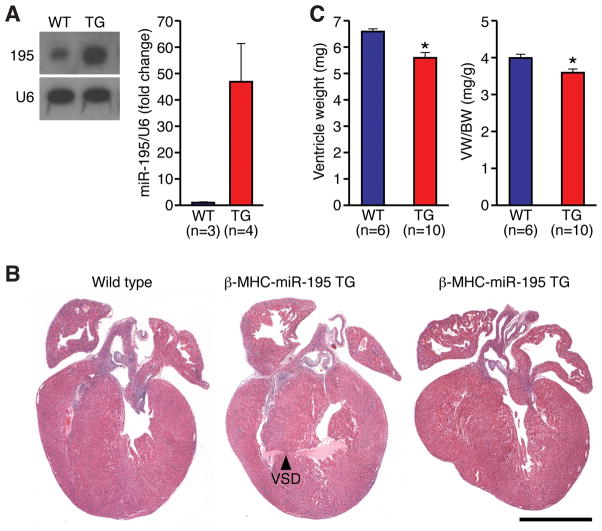
In order to determine whether up-regulation of the miR-15 family plays a role in arresting cardiomyocyte proliferation, we generated transgenic mice over-expressing miR-195 in the embryonic heart under the control of the β-myosin heavy chain (βMHC) promoter. Because over-expression of miR-195 caused perinatal cardiomyopathy and early lethality in two independent transgenic (Tg) founders (Online Figure III), we focused our analyses on a third Tg line, which was viable. We were unable to obtain additional viable lines of βMHC-miR-195 transgenic mice, despite multiple rounds of pronuclear injection, suggesting that expression of miR-195 above a threshold is lethal, which most likely reflects the potent anti-proliferative activity of this miRNA (see below). Northern blot and real-time PCR analysis showed miR-195 to be over-expressed in Tg cardiac ventricles ~45-fold above wild-type (WT) levels (Figure 2A). Histological analysis of Tg hearts between P1–P3 revealed that mice over-expressing miR-195 developed congenital heart defects including ventricular septal defects (4/18 analyzed, Figure 2B) and ventricular hypoplasia, which appeared to predominantly affect the right ventricle (2/18 analyzed, Figure 2B). On average, neonatal Tg hearts displayed a significant reduction in ventricle weight (~16%) and ventricle weight to body weight ratio (~10%) at P1, compared to WT littermates (Figure 2C). Interestingly, Tg mice developed a slow-onset cardiomyopathy and died prematurely (Online Figure IVA), beginning at 5–6 months of age, with evidence of cardiac enlargement and dilation apparent upon necropsy (Online Figure IVB). Since we did not observe ventricular septal defects (VSDs) in βMHC-miR-195 Tg mice that survived to adulthood, we assume that these VSDs either spontaneously closed during early post-natal life or that these mice died soon after birth.
Figure 2. Cardiac-specific over-expression of miR-195 during embryogenesis is associated with a spectrum of cardiac malformations and reduced heart size at birth.
A, Northern blot (Left) and real-time PCR (Right) analysis of miR-195 expression levels inβMHC-miR-195 transgenic hearts at P1. B, H&E-stained sections show that miR-195 over-expression causes congenital cardiac malformations. Three sections are shown from 1 day-old wild-type (Left) and βMHC-miR-195 transgenic (Middle and Right) neonatal mice. Note large apical ventricular septal defect and ventricular hypoplasia in transgenic hearts (Middle and Right, respectively). Scale bar = 1mm. C, Ventricle weight and ventricle weight to body weight ratio for wild-type (WT) and βMHC-miR-195 transgenic (TG) mice at P1. Values presented as mean±SEM, n=6–10 per group (as indicated); *P<0.05.
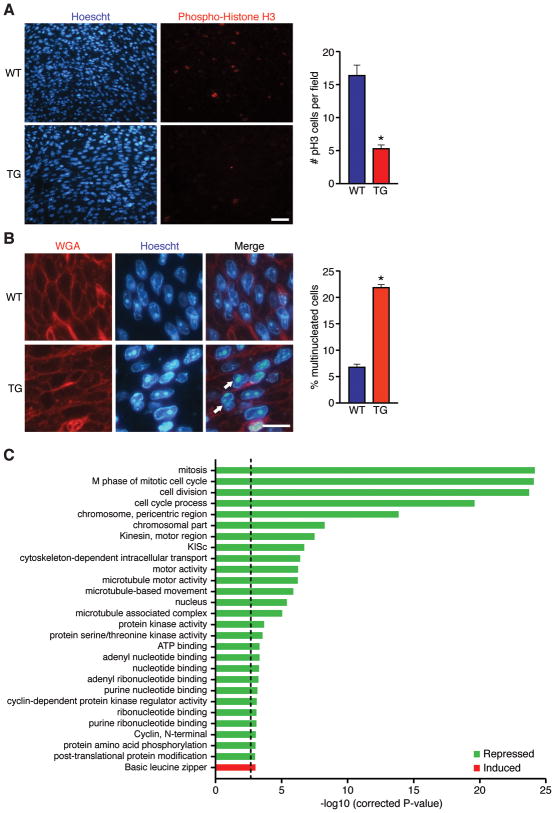
βMHC-miR-195 transgenic hearts were examined at P1 for evidence of defective mitosis, increased apoptosis or reduced cell size, all of which could potentially explain why these hearts are smaller at birth. We did not detect any significant differences in cell size or apoptosis between Tg hearts and WT littermates at P1 (Online Figure V). In contrast, a marked reduction (~3-fold) in the number of cells undergoing mitosis and an increased proportion of multinucleated myocytes (~3-fold) were noted in Tg hearts at P1 (Figure 3A and B). These data indicate that over-expression of miR-195 in vivo inhibits cardiomyocyte mitotic progression and induces premature cell cycle arrest.
Figure 3. Over-expression of miR-195 impairs cardiomyocyte proliferation and represses mitotic gene expression.
A, Sections from 1 day-old wild-type (WT) and transgenic (TG) hearts stained with phospho-Histone H3 (pH3) and Hoechstt 33342 to label mitotic cells and total nuclei, respectively. Scale bar = 40 μm. Quantification of pH3 staining in WT and TG hearts at P1 is shown (Right). Values presented as mean±SEM; *P<0.05. B, Sections from 1 day-old WT and TG hearts stained with wheat germ agglutinin (WGA) and Hoechst 33342 to label cell membranes and nuclei, respectively. Arrows denote multinucleated cells. Scale bar = 20 μm. Quantification of the percentage of multinucleated cells in WT and TG hearts at P1 is shown (Right). Values presented as mean±SEM, *P<0.05. C, Ontology analysis of genes differentially regulated in miR-195 transgenic hearts at P1. Genes that were either repressed (green) or induced (red) by miR-195 over-expression were clustered on the basis of shared ontology using the DAVID algorithm. A corrected P-value less than 0.001 was considered significant.
To test the direct effects of miR-195 on cardiomyocyte proliferation, we infected cultured neonatal rat cardiomyocytes with an adenovirus expressing miR-195 (Ad195).17 miR-195 expression levels were titrated to match endogenous levels of miR-195 in the P14 heart (Online Figure VIA). Over-expression of miR-195 in cultured neonatal cardiomyocytes was associated with cellular hypertrophy (Online Figure VIB), as previously reported.17 In addition, we found that a greater proportion of neonatal cardiomyocytes over-expressing miR-195 were binucleate (Online Figure VI, B and C). Cell cycle profile analysis indicated that neonatal cardiomyocytes expressing miR-195 accumulated in G2 phase (Online Figure VID), consistent with a block in G2/M phase transition and defective mitotic progression. Similar results were obtained in the proliferative myoblastic cell line, H9c2, which also displayed an accumulation of cells in G2 phase following over-expression of miR-195 (Online Figure VIE).
miR-195 Negatively Regulates the Expression of Cell Cycle Genes
Global changes in the transcriptional profile of neonatal hearts over-expressing miR-195 were assessed by microarray. We identified 228 genes that were down-regulated in Tg cardiac ventricles at P1 and 100 genes that were up-regulated at this time point (Online Figure VIIA). Among the transcripts that were significantly repressed in Tg ventricles at P1, gene ontology cluster analysis revealed a striking enrichment for genes involved in mitosis and various aspects of cell division (Figure 3C). Thirty-nine of the genes that were down-regulated in Tg ventricles (~17%) contained predicted miR-15 family binding sites within their 3’UTRs, but there was little overlap between different target prediction algorithms (Online Figure VIIB).
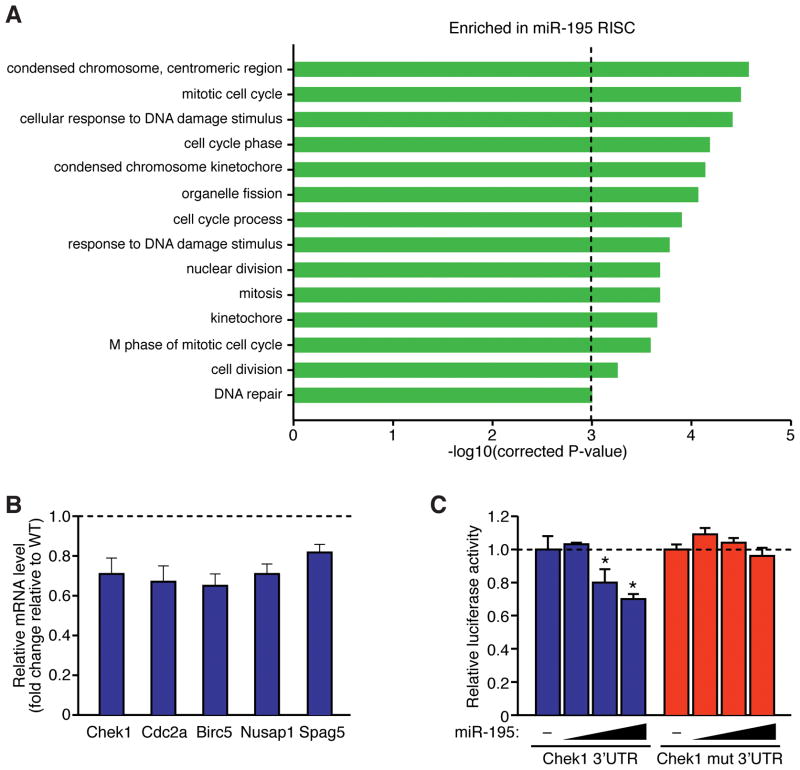
In an attempt to identify direct in vivo targets of miR-195, we employed a recently described method for context-specific detection of miRNA targets by RNA-induced silencing complex RNA sequencing (RISC-seq).19 Using next generation massively parallel sequencing of unamplified mRNAs isolated from Argonaute 2 (Ago2) immunoprecipitates from Tg and WT cardiac ventricles at P1, we were able to identify a number of transcripts enriched (>2-fold) within the miR-195 RISCome (Online Figure VII, C and D). Gene ontology cluster analysis of transcripts that were over-represented within the miR-195 RISCome demonstrated significant enrichment for genes involved in the regulation of the mitotic cell cycle, chromosome condensation and the cellular response to DNA damage (Figure 4A). Among the 547 genes enriched (>2-fold) within the miR-195 RISCome, 128 genes (~23%) contained predicted miR-15 family binding sites within their 3’UTRs. Following DAVID gene ontology cluster analysis, we identified 20 genes that were enriched within the miR-195 RISCome that are known regulators of the mitotic cell cycle (Online Figure VIIIA). While five of these mitotic genes (Chek1, Cdc2a, Birc5, Nusap1 and Spag5) contained predicted miR-15 family binding sites within their 3’UTRs (Online Figure VIIIA), the checkpoint kinase 1 (Chek1) binding site was the only one conserved from mice to humans (Online Figure VIII, A and B). Furthermore, real-time PCR analysis indicated that Chek1, Cdc2a, Birc5, Nusap1 and Spag5 were all robustly down-regulated in the heart between P7 and P14, which was inversely correlated with the endogenous expression pattern of miR-195 (Online Figure IX and data not shown). Real-time PCR analysis demonstrated an ~20–40% reduction in the expression levels of Chek1, Cdc2a, Birc5, Nusap1 and Spag5 in βMHC-miR-195 Tg hearts at P1 (Figure 4B), indicating that miR-195 modestly represses the expression of these cell cycle genes in vivo.
Figure 4. RISC-sequencing of miR-195 transgenic hearts identifies several target genes implicated in cell cycle regulation.
A, Ontology analysis of genes enriched in miR-195 RISC. Genes that were enriched (>2-fold) in the RISC of neonatal miR-195 transgenic hearts were clustered on the basis of shared ontology using the DAVID algorithm. A corrected P-value less than 0.001 was considered significant. B, Real-time PCR analysis for genes with predicted miR-15 binding sites in transgenic and wild-type hearts at P1. Expression levels of each gene are presented as a fold change relative to WT. C, Luciferase assay demonstrating the response of wild-type Chek1 3’ UTR, or of the same DNA fragment containing a mutation of the miR-15 family binding site, to increasing amounts of miR-195. A total of 0, 1, 100 or 1000 nM of miR-195 mimic was cotransfected with the luciferase reporter construct in COS cells. Values presented as mean±SEM; n=3 per group, *P<0.05.
We next conducted 3’UTR luciferase reporter assays to determine whether miR-195 could directly target Chek1 for repression. A plasmid consisting of the Chek1 3’UTR linked to a constitutively active luciferase reporter was transfected into COS cells along with increasing amounts of miR-195 mimic. The Chek1 3’UTR luciferase reporter displayed a dose-dependent repression in activity by miR-195 (Figure 4C). Mutation of the miR-195 target site abolished repression by miR-195, demonstrating that this effect was dependent upon binding of miR-195 to its target site within the 3’UTR of Chek1 (Figure 4C).
Post-natal Inhibition of the miR-15 Family In Vivo Induces Cardiomyocyte Mitotic Entry
To further interrogate the potential role of the miR-15 family in suppression of cardiomyocyte proliferation, we knocked down the miR-15 family in vivo in neonatal mice, using locked-nucleic acid (LNA)-modified oligonucleotides directed against miR-15b and miR-16 (Online Table II). Control mice received an oligonucleotide directed against a miRNA specific for C. elegans that is not present in mammals. In order to prevent post-natal up-regulation of the miR-15 family, neonatal mice received antimiRs (25 mg/kg) directed against the miR-15 family members, miR-15b (Anti-15) and miR-16 (Anti-16), via subcutaneous injection once per day from P2-P4 (Figure 5A). Initial pilot studies indicated that administration of either Anti-15 or Anti-16 alone was not sufficient to knock-down expression of all miR-15 family members in the heart (Online Figure XA). However, when Anti-15 and Anti-16 (Anti15/16) were administered in combination (25 mg/kg each), we observed significant repression of all miR-15 family members in the heart at P12 (i.e. 10 days following the first treatment) (Online Figure IXA). Therefore, experimental animals received a combination of Anti-15/16, delivered via subcutaneous injection between P2-P4, and were harvested at P12 for confirmation of miRNA knockdown and assessment of target regulation and cardiomyocyte proliferation (Figure 5A). Successful knockdown of all miR-15 family members was achieved using this strategy and documented by Northern blot (Online Figure XB) and quantified by real-time PCR (Figure 5B).
Figure 5. Post-natal inhibition of the miR-15 family induces cardiomyocytes to enter mitosis.
A, Schematic of the antimiR experiment designed to knock-down miR-15 family expression during early post-natal life. B, Real-time PCR analysis of miR-15 family member expression in the heart at P12, following administration of miR15/16 antimiRs at P2–P4. miR expression levels normalized to U6 and presented as a fold change relative to mice treated with a control oligo. Values presented as mean±SEM, n=6 per group, *P<0.05. C, Sections from P12 control and anti-15/16-treated hearts stained with phospho-histone H3 (green) and cardiac troponin T (red) to label mitotic cardiomyocytes (arrow). Scale bar = 20 μm. Quantification of the number of pH3 cardiomyocytes (per field) in WT and TG hearts at P1 is shown (Right). Data presented as cells/field for n=3 independent samples per group (10 fields of cells assessed per heart). Values presented as mean±SEM; *P<0.05. D, Real-time PCR analysis of Chek1 expression following antimiR-mediated knock-down of miR-15 family members in the neonatal heart. Expression levels shown as a fold change relative to mice treated with a control oligo. Values presented as mean±SEM, n = 6 per group; *P<0.05.
Post-natal inhibition of miR-15 family function was associated with an increased number of cardiomyocytes that stained positive for phospho-histone H3 (Figure 5C and D), as well as a higher number of cardiomyocytes displaying disorganized sarcomeric structures (Online Figure XI, A and B), characteristic of cardiomyocytes undergoing mitosis.22 Cardiomyocyte size was unaffected by post-natal knockdown of the miR-15 family (Online Figure XI, C and D). Importantly, three out of five predicted miR-15 family targets from our RISC-seq studies were enhanced by miR-15 family knockdown, including the highly conserved target Chek1 (Figure 5E), as well as the non-conserved targets Cdc2a and Spag5 (Online Figure XII). While these data imply that post-natal inhibition of the miR-15 family increases cardiomyocyte mitotic entry and progression, we detected very few cardiomyocytes staining positive for Aurora B kinase at P12 in control and Anti-15/16-treated groups (data not shown), suggesting that loss of miR-15 family function was not sufficient to drive complete mitotic progression through cytokinesis. Taken together, our data suggest that post-natal up-regulation of the miR-15 family may represent an important regulatory brake on the cardiomyocyte mitotic machinery, which contributes to post-natal cell cycle withdrawal.
Discussion
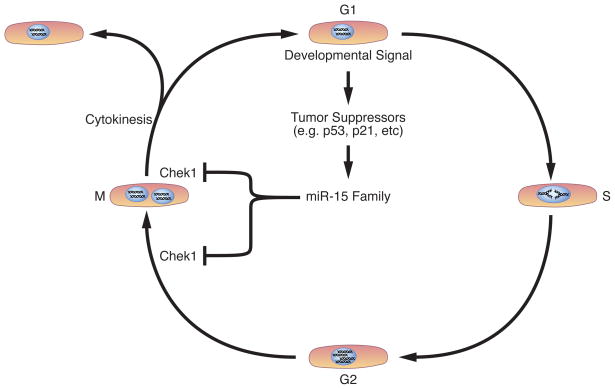
Here, we describe a group of miRNAs belonging to the miR-15 family that are up-regulated after birth and which contribute to cardiomyocyte mitotic arrest during the neonatal period. Over-expression of miR-195 in the developing heart is associated with a spectrum of congenital cardiac abnormalities and premature cell cycle arrest. We provide evidence that miR-195 regulates a number of cell cycle genes in vivo, including Chek1. Post-natal inhibition of the miR-15 family in neonatal mice is associated with an increased number of mitotic cardiomyocytes and elevated expression levels of Chek1. The current study identifies a novel role for miRNAs in regulating cardiomyocyte mitotic arrest during early post-natal life and a proposed model to account for the role of the miR-15 family in this process is outlined in Figure 6.
Figure 6.
Proposed model for the regulation of cardiomyocyte proliferation by the miR-15 family. The miR-15 family is proposed to lie downstream of a tumor suppressor pathway that is activated in the heart shortly after birth. MiR-15 family members repress cardiomyocyte proliferation by inhibiting a number of cell cycle genes involved in G2/M phase transition and mitotic progression, including Chek1.
Our data support previous studies in which miR-15 family members were found to inhibit cell proliferation. The miR-15a/16-1 cluster is located within the chromosomal region 13q14, which is commonly deleted or down-regulated in chronic lymphocytic leukemia (CLL).23 Genetic deletion of miR-15a/16-1 in the mouse accelerates proliferation of B cells and contributes to the development of CLL.24 The miR-15 family also appears to repress cell proliferation in a number of non-lymphoid cell systems and deletion or down-regulation of miR-15 family members is associated with the pathogenesis of a number of solid tumors.25–28 The current study is the first to implicate the miR-15 family in the physiological regulation of the cardiac cell cycle. Although the role of miR-15 family members in the developmental regulation of post-natal proliferative growth arrest in other tissues has not been assessed, it is probable that the miR-15 family also exerts tumor suppressor activity in multiple adult tissues. Given that the systemic antimiR administration protocol used in the current study is likely to have effects on multiple organ systems, it will be interesting to examine the relative myocyte-autonomous and non-autonomous effects of the miR-15 family in the future.
A plethora of proliferation-associated genes has been reported to be targeted by the miR-15 family.24–28 We used a non-biased approach for in vivo identification of miR-15 family target genes in the heart of neonatal miR-195 transgenic mice using RISC-seq.19 Several mitotic genes were enriched within the miR-195 RISCome, including a number of genes containing predicted miR-15 family binding sites located within their 3’UTRs, but Chek1 was the only predicted cell cycle-associated gene with a miR-15 family binding site in its 3’UTR that is conserved in humans. Interestingly, many genes that were enriched within the miR-195 RISCome did not contain predicted miR-15 family binding sites within their 3’UTR, which could reflect potential binding to sites located within the 5’UTR, coding sequence or introns of target transcripts,29 or possibly secondary effects of miRNA over-expression. Future studies employing RISC-seq approaches in combination with mRNA footprinting29 will likely shed further light on the precise mechanisms of miRNA target recognition in vivo.
The current findings are consistent with earlier observations in mice lacking miR-15a/16-1, in which Chek1 levels were found to be elevated.24 Chek1 plays a crucial role in the prevention of genomic instability and mediates the cellular response to DNA replication errors, DNA damage and chromatin breaks.30 Furthermore, Chek1 has been implicated in the regulation of mitosis and coordinates progression through the G2/M and spindle checkpoints, as well as chromosome segregation and cytokinesis.30, 31 Of particular relevance to the current findings, Chek1 has been shown to translocate to the spindle midzone during anaphase and later accumulates at the cytokinetic midbody where it is thought to regulate the Aurora B-mediated abscission checkpoint.30, 31 Heterozygous genetic deletion of Chek1 in mouse primary mammary epithelial cells is linked with increased chromosome lagging, cytokinetic regression and binucleation.31 Thus, the developmental inactivation of Chek1 in the heart after birth, which we show is at least in part mediated by post-natal induction of the miR-15 family, may contribute to the onset of cardiomyocyte binucleation during the neonatal period.
This is the first study to implicate the miR-15 family in heart development. Of particular interest, over-expression of miR-195 in the developing heart is associated with a number of congenital heart abnormalities including ventricular septal defects and severe ventricular hypoplasia. Ventricular septal defects and ventricular hypoplasia are commonly observed in mouse models with cardiomyocyte proliferative defects.12, 32, 33 Moreover, a number of cell cycle genes are dysregulated in human patients with ventricular septal defects and hypoplastic heart syndrome.34, 35 The current findings suggest that fine tuning of cardiac cell cycle genes by miRNAs may be an important regulatory component of cardiac development and congenital heart disease.
We previously reported that post-natal over-expression of miR-195 in the heart under the control of the αMHC promoter resulted in pathological cardiac remodeling and heart failure.17 Interestingly, the current study suggests that embryonic over-expression of miR-195 in the heart can also drive adult cardiac hypertrophy and dilation, as well as premature death. It is possible that the phenotype observed in αMHC-miR-195 Tg mice also has developmental origins, as the αMHC transgene is turned on perinatally.36 Indeed, premature cardiomyocyte binucleation and cardiac growth restriction during early life have been reported to precede the onset of pathological cardiac hypertrophy in several experimental models of heart disease.37–39 Although a number of studies have identified adult cardiac pathologies in mice with congenital heart abnormalities,12, 40 the potential contribution of developmental defects related to cell cycle control in the pathogenesis of adult human cardiomyopathies is not well understood.
The miR-15 family has been recently shown to lie downstream of the p53 tumor suppressor pathway.41–43 Although the upstream signals for postnatal induction of the miR-15 family in the heart are currently unknown, it is tempting to speculate that the miR-15 family is an important downstream effector of a tumor suppressor network that is activated in the heart shortly after birth (Figure 6). Further analysis of the cis regulatory elements and upstream signals responsible for post-natal up-regulation of the miR-15 family may shed light on the molecular determinants of cardiomyocyte mitotic arrest and open new avenues to cardiomyocyte cell cycle re-induction for cardiac repair.
Supplementary Material
Novelty and Significance.
What is Known?
The mammalian heart can regenerate following injury for a short period after birth, but this regenerative potential is lost as cardiac myocytes mature and lose their ability to divide.
The regulatory mechanisms that govern cardiac myocyte cell cycle withdrawal during early post-natal development are poorly understood, but appear to involve the coordinated repression of a large number of cell cycle genes.
microRNAs are small noncoding RNAs that inhibit the expression of large numbers of target mRNAs, but their role in cardiac myocyte cell cycle regulation during neonatal development has not been explored.
What New Information Does This Article Contribute?
Multiple members of the miR-15 family are up-regulated post-natally in the heart during the time window when cardiac myocytes exit the cell cycle.
Over-expression of miR-195 (a member of the miR-15 family) in the embryonic heart is associated with a number of congenital heart defects including ventricular septal defects and cardiac myocyte hypoplasia.
The miR-15 family regulates post-natal cardiac myocyte cell cycle arrest by repressing the expression of multiple cell cycle genes in the heart.
We have recently demonstrated that the neonatal mammalian heart can regenerate following injury and that this regenerative potential is lost as cardiac myocytes mature and lose their ability to divide. The regulatory mechanisms that govern post-natal cardiac myocyte cell cycle withdrawal are poorly understood. Given the vast regulatory potential of microRNAs, which often inhibit the expression of large numbers of target mRNAs, we searched for microRNAs involved in cardiac myocyte cell cycle arrest. Here, we report that multiple members of the miR-15 family are up-regulated in the heart shortly after birth. We demonstrate that over-expression of miR-195 (a member of the miR-15 family) in the embryonic heart is associated with a spectrum of congenital cardiac abnormalities including ventricular septal defects and cardiac hypoplasia. Furthermore, using gain- and loss-of-function approaches in the mouse, coupled with comprehensive and unbiased analyses of microRNA targets in vivo, we show that the miR-15 family inhibits cardiac myocyte proliferation by repressing the expression of multiple cell cycle genes. For the first time, these studies implicate the miR-15 family in the regulation of the cardiac cell cycle. This knowledge may facilitate the future development of therapeutic strategies for the induction of cardiac myocyte proliferation and ultimately regeneration of the adult heart.
Acknowledgments
We gratefully acknowledge miRagen Therapeutics for providing LNA-modified antimiRs, John McAnally for generation of transgenic mice, Jimin Pei for assistance with bioinformatics, James Richardson for assistance with pathology, John Shelton for technical assistance and Jose Cabrera for graphical assistance. We thank Hesham Sadek and Robert Frost for helpful discussions.
Sources of Funding
Work in the laboratory of E.N.O. was supported by grants from the NIH, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Foundation Leducq’s Transatlantic Network of Excellence in Cardiovascular Research Program, the American Heart Association – Jon Holden deHaan Foundation, and the Robert A. Welch Foundation. E.R.P. was supported by an overseas postdoctoral fellowship from the National Health and Medical Research Council and National Heart Foundation of Australia.
Non-standard Abbreviations and Acronyms
- Ad195
adenovirus expressing miR-195
- AdGal
adenovirus expressing β-galactosidase
- Ago2
Argonaute 2
- Anti-15/16
antimiRs directed against miR-15b and miR-16
- βMHC
beta myosin heavy chain
- Chek1
Checkpoint kinase 1
- CLL
chronic lymphocytic leukemia
- E
embryonic day
- LNA
locked nucleic acid
- miRNA
microRNA
- P
post-natal day
- RISC
RNA-induced silencing complex
- RISCome
RNA-induced silencing complex transcriptome
- RNA-seq
RNA-induced silencing complex (RISC) RNA sequencing
- Tg
transgenic
- UTR
untranslated region
- WT
wild-type
Footnotes
Disclosures
E.N.O holds equity in miRagen Therapeutics, a company focused on developing miRNA-based therapies for cardiovascular disease.
References
- 1.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- 3.Walsh S, Ponten A, Fleischmann BK, Jovinge S. Cardiomyocyte cell cycle control and growth estimation in vivo--an analysis based on cardiomyocyte nuclei. Cardiovasc Res. 2010;86:365–373. doi: 10.1093/cvr/cvq005. [DOI] [PubMed] [Google Scholar]
- 4.Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle regulation. Circ Res. 2002;90:1044–1054. doi: 10.1161/01.res.0000020201.44772.67. [DOI] [PubMed] [Google Scholar]
- 7.Bersell K, Arab S, Haring B, Kuhn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138:257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 8.Engel FB, Hsieh PC, Lee RT, Keating MT. FGF1/p38 MAP kinase inhibitor therapy induces cardiomyocyte mitosis, reduces scarring, and rescues function after myocardial infarction. Proc Natl Acad Sci U S A. 2006;103:15546–15551. doi: 10.1073/pnas.0607382103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuhn B, del Monte F, Hajjar RJ, Chang YS, Lebeche D, Arab S, Keating MT. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13:962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 10.Ahuja P, Sdek P, MacLellan WR. Cardiac myocyte cell cycle control in development, disease, and regeneration. Physiol Rev. 2007;87:521–544. doi: 10.1152/physrev.00032.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 14.Small EM, Frost RJ, Olson EN. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rindt H, Gulick J, Knotts S, Neumann J, Robbins J. In vivo analysis of the murine beta-myosin heavy chain gene promoter. J Biol Chem. 1993;268:5332–5338. [PubMed] [Google Scholar]
- 16.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, Childs G, Harris S, Owens GK, Olson EN. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–2636. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd RISC RNA sequencing for context-specific identification of in vivo microRNA targets. Circ Res. 2011;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lompre AM, Nadal-Ginard B, Mahdavi V. Expression of the cardiac ventricular alpha-and beta-myosin heavy chain genes is developmentally and hormonally regulated. J Biol Chem. 1984;259:6437–6446. [PubMed] [Google Scholar]
- 22.Ahuja P, Perriard E, Perriard JC, Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J Cell Sci. 2004;117:3295–3306. doi: 10.1242/jcs.01159. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Bandi N, Zbinden S, Gugger M, Arnold M, Kocher V, Hasan L, Kappeler A, Brunner T, Vassella E. miR-15a and miR-16 are implicated in cell cycle regulation in a Rb-dependent manner and are frequently deleted or down-regulated in non-small cell lung cancer. Cancer Res. 2009;69:5553–5559. doi: 10.1158/0008-5472.CAN-08-4277. [DOI] [PubMed] [Google Scholar]
- 26.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D'Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 27.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Fu H, Sun F, Zhang H, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. miR-16 family induces cell cycle arrest by regulating multiple cell cycle genes. Nucleic Acids Res. 2008;36:5391–5404. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peddibhotla S, Rosen JM. Chking and executing cell division to prevent genomic instability. Cell Cycle. 2009;8:2339–2342. doi: 10.4161/cc.8.15.9169. [DOI] [PubMed] [Google Scholar]
- 31.Peddibhotla S, Lam MH, Gonzalez-Rimbau M, Rosen JM. The DNA-damage effector checkpoint kinase 1 is essential for chromosome segregation and cytokinesis. Proc Natl Acad Sci U S A. 2009;106:5159–5164. doi: 10.1073/pnas.0806671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King JC, Moskowitz IP, Burgon PG, Ahmad F, Stone JR, Seidman JG, Lees JA. E2F3 plays an essential role in cardiac development and function. Cell Cycle. 2008;7:3775–3780. doi: 10.4161/cc.7.23.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gambetta K, Al-Ahdab MK, Ilbawi MN, Hassaniya N, Gupta M. Transcription repression and blocks in cell cycle progression in hypoplastic left heart syndrome. Am J Physiol Heart Circ Physiol. 2008;294:H2268–2275. doi: 10.1152/ajpheart.91494.2007. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Zhou L, Yang R, Sheng Y, Sun W, Kong X, Cao K. Identification of differentially expressed genes in human heart with ventricular septal defect using suppression subtractive hybridization. Biochem Biophys Res Commun. 2006;342:135–144. doi: 10.1016/j.bbrc.2006.01.113. [DOI] [PubMed] [Google Scholar]
- 36.Ng WA, Grupp IL, Subramaniam A, Robbins J. Cardiac myosin heavy chain mRNA expression and myocardial function in the mouse heart. Circ Res. 1991;68:1742–1750. doi: 10.1161/01.res.68.6.1742. [DOI] [PubMed] [Google Scholar]
- 37.Levkau B, Schafers M, Wohlschlaeger J, von Wnuck Lipinski K, Keul P, Hermann S, Kawaguchi N, Kirchhof P, Fabritz L, Stypmann J, Stegger L, Flogel U, Schrader J, Fischer J, Hsieh P, Ou YL, Mehrhof F, Tiemann K, Ghanem A, Matus M, Neumann J, Heusch G, Schmid KW, Conway EM, Baba HA. Survivin determines cardiac function by controlling total cardiomyocyte number. Circulation. 2008;117:1583–1593. doi: 10.1161/CIRCULATIONAHA.107.734160. [DOI] [PubMed] [Google Scholar]
- 38.Porrello ER, Bell JR, Schertzer JD, Curl CL, McMullen JR, Mellor KM, Ritchie RH, Lynch GS, Harrap SB, Thomas WG, Delbridge LM. Heritable pathologic cardiac hypertrophy in adulthood is preceded by neonatal cardiac growth restriction. Am J Physiol Regul Integr Comp Physiol. 2009;296:R672–680. doi: 10.1152/ajpregu.90919.2008. [DOI] [PubMed] [Google Scholar]
- 39.Porrello ER, Widdop RE, Delbridge LM. Early origins of cardiac hypertrophy: does cardiomyocyte attrition programme for pathological 'catch-up' growth of the heart? Clin Exp Pharmacol Physiol. 2008;35:1358–1364. doi: 10.1111/j.1440-1681.2008.05036.x. [DOI] [PubMed] [Google Scholar]
- 40.Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, Elliott DE, Prall OW, Black BL, Fatkin D, Harvey RP. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- 41.Boominathan L. The tumor suppressors p53, p63, and p73 are regulators of microRNA processing complex. PLoS One. 2010;5:e10615. doi: 10.1371/journal.pone.0010615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, Goldstein I, Madar S, Goldfinger N, Borresen-Dale AL, Ginsberg D, Harris CC, Pilpel Y, Oren M, Rotter V. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol Syst Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature. 2009;460:529–533. doi: 10.1038/nature08199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.