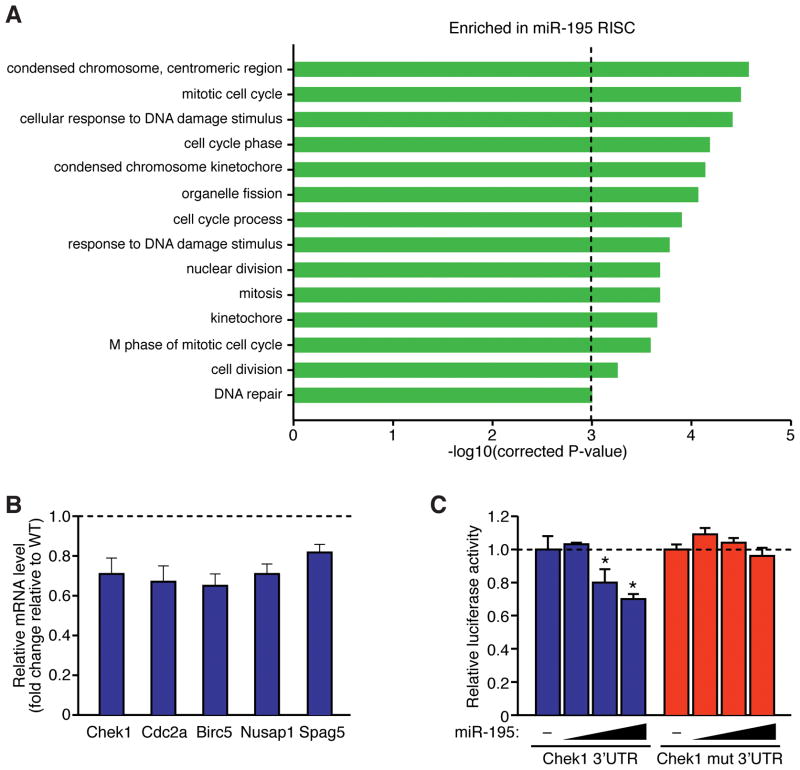
Figure 4. RISC-sequencing of miR-195 transgenic hearts identifies several target genes implicated in cell cycle regulation.
A, Ontology analysis of genes enriched in miR-195 RISC. Genes that were enriched (>2-fold) in the RISC of neonatal miR-195 transgenic hearts were clustered on the basis of shared ontology using the DAVID algorithm. A corrected P-value less than 0.001 was considered significant. B, Real-time PCR analysis for genes with predicted miR-15 binding sites in transgenic and wild-type hearts at P1. Expression levels of each gene are presented as a fold change relative to WT. C, Luciferase assay demonstrating the response of wild-type Chek1 3’ UTR, or of the same DNA fragment containing a mutation of the miR-15 family binding site, to increasing amounts of miR-195. A total of 0, 1, 100 or 1000 nM of miR-195 mimic was cotransfected with the luciferase reporter construct in COS cells. Values presented as mean±SEM; n=3 per group, *P<0.05.