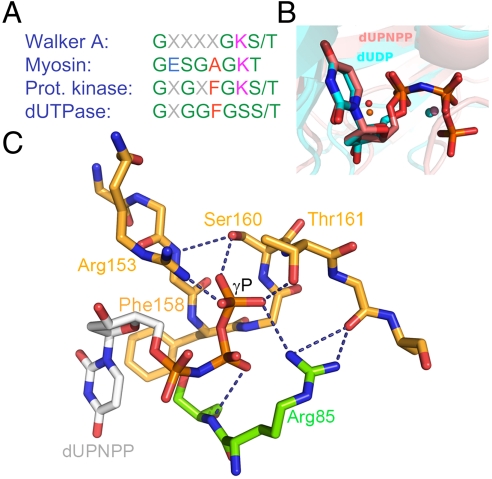
Fig. 1.
The P-loop-like C-terminal motif V and its interactions with the substrate, (A), Consensus P-loop motifs from well known P-loop NTPases compared with the P-loop-like sequence of dUTPases. Coloring is as follows, polar residues, green; apolar r., red; negatively charged r., blue; and positively charged r., magenta (B), Alignment of two dUTPase active sites containing uridine di- and triphosphate ligands [atomic coloring with cyan and pink carbons, PDB IDs 1SLH (9) and 2HQU (8) from M. tuberculosis and Homo sapiens, respectively]. The positions of the α-β phosphates are identical and the catalytic water (orange for the tri- and magenta for the diphosphate structure) and Mg2+ ions (spheres in colors corresponding to the related nucleosides) are in place for putative hydrolysis of the α-β phosphate bond in both structures. (C), Coordination of the γ-P of the substrate by amino acids subjected to mutation in this study. The homotrimeric protein (2, PDB ID: 2HQU) is colored by subunits (subunit C, atomic coloring with orange carbons; subunit B atomic coloring with green carbons). dUPNPP is depicted as atomic colored sticks with gray carbons. Dashed lines depict hydrogen bonds partially abolished by the mutations thus hampering the coordination of the γ-P. Arg85 of subunit B is shown to complete the H-bonding network around the γ-P. Phe158, which engages in an aromatic stacking with uracil, is exchanged to a Trp to yield the specific fluorescence signal throughout this study. For better viewing, these structural images are also shown in stereo mode in Fig. S1.