Figure 5.
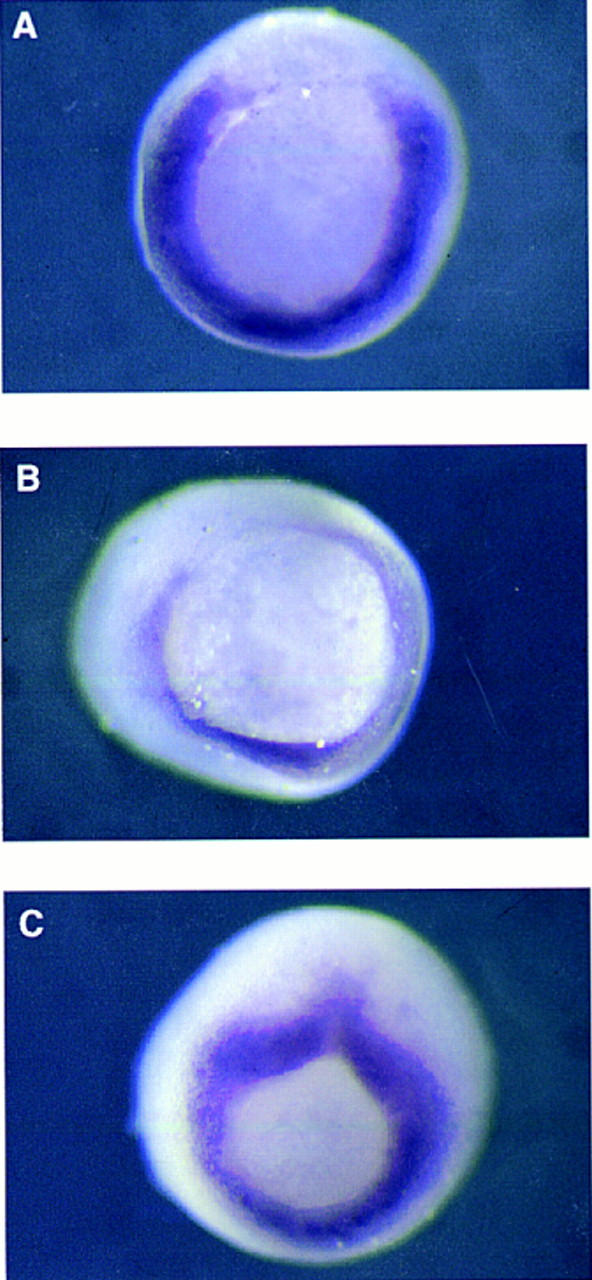
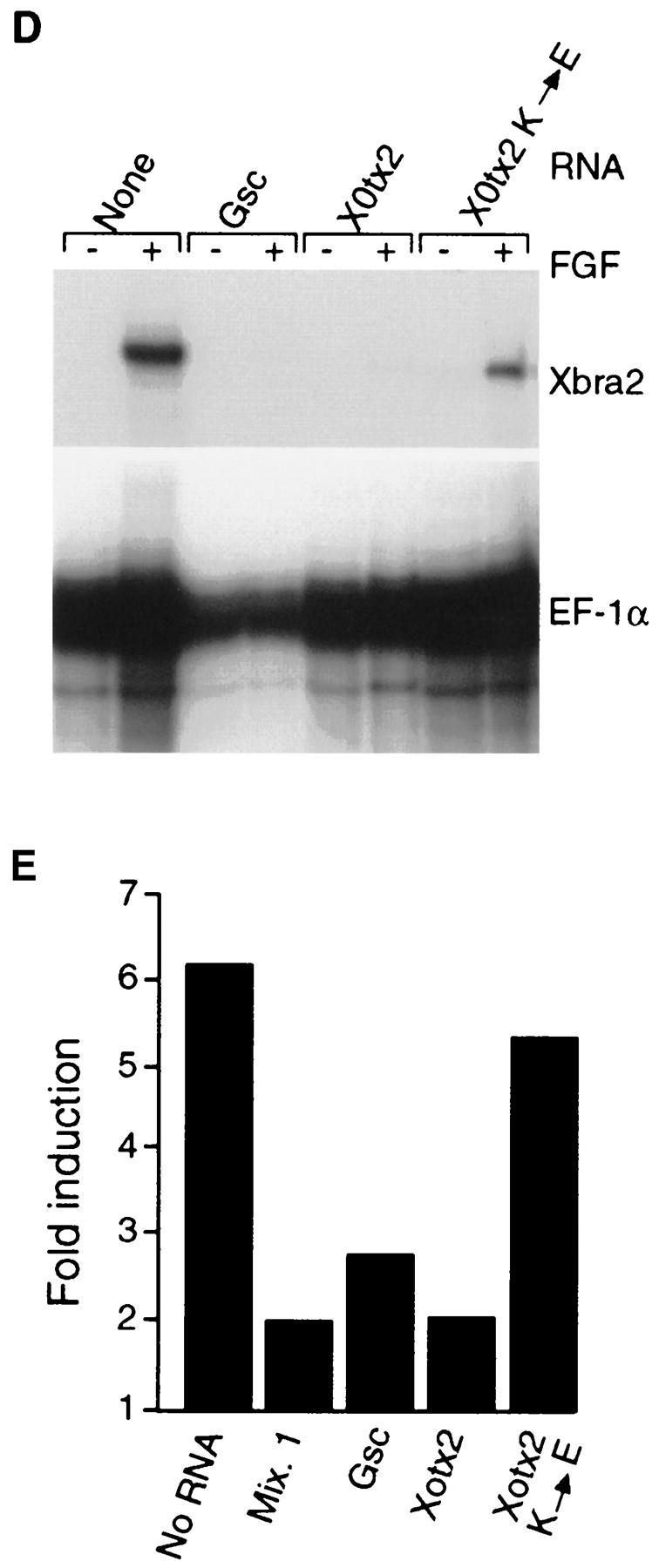
Suppression of Xbra expression in gastrula-stage embryos by paired-type homeodomain proteins. Embryos at the four-cell stage were injected equatorially with 500 pg of goosecoid (A), Xotx2 (B), or Xotx2 K → E (C) RNA in one blastomere, and the expression pattern of Xbra was analyzed by whole-mount in situ hybridization at the early gastrula stage. Note lack of Xbra expression in part of the equatorial region of embryos in A and B. All embryos shown are from the same experiment; that in C is at a slightly later stage than those in A and B. (D) Suppression of FGF-induced Xbra2 mRNA in animal caps by goosecoid, Mix.1, and Xotx2 but not by Xotx2 K → E. Embryos at the one-cell stage were injected with 1 ng of the indicated RNAs, and animal caps were cut at stage 8 and incubated in FGF until stage 12. Xbra2 and EF-1α transcripts were detected by RNase protection. Note that FGF-induced expression of Xbra2 is strongly inhibited by goosecoid, Mix.1, and Xotx2 but not by Xotx2 K → E. (E) Suppression of FGF-induced Xbra2.pGL2 luciferase activity in animal caps by goosecoid, Mix.1, and Xotx2 but not by Xotx2 K → E. Embryos were coinjected with 1 ng of the indicated RNAs, 30 pg of Xbra2.pGL2, and 10 pg of pRL–SV reference plasmid. Animal caps were excised at stage 8 and incubated with FGF until stage 12.5, and firefly and Renilla luciferase activities were determined. Activity of Xbra2.pGL2 is presented as fold activation over uninduced levels. This experiment was repeated twice for all four RNAs and four additional times with goosecoid, with similar results.
