Figure 7.
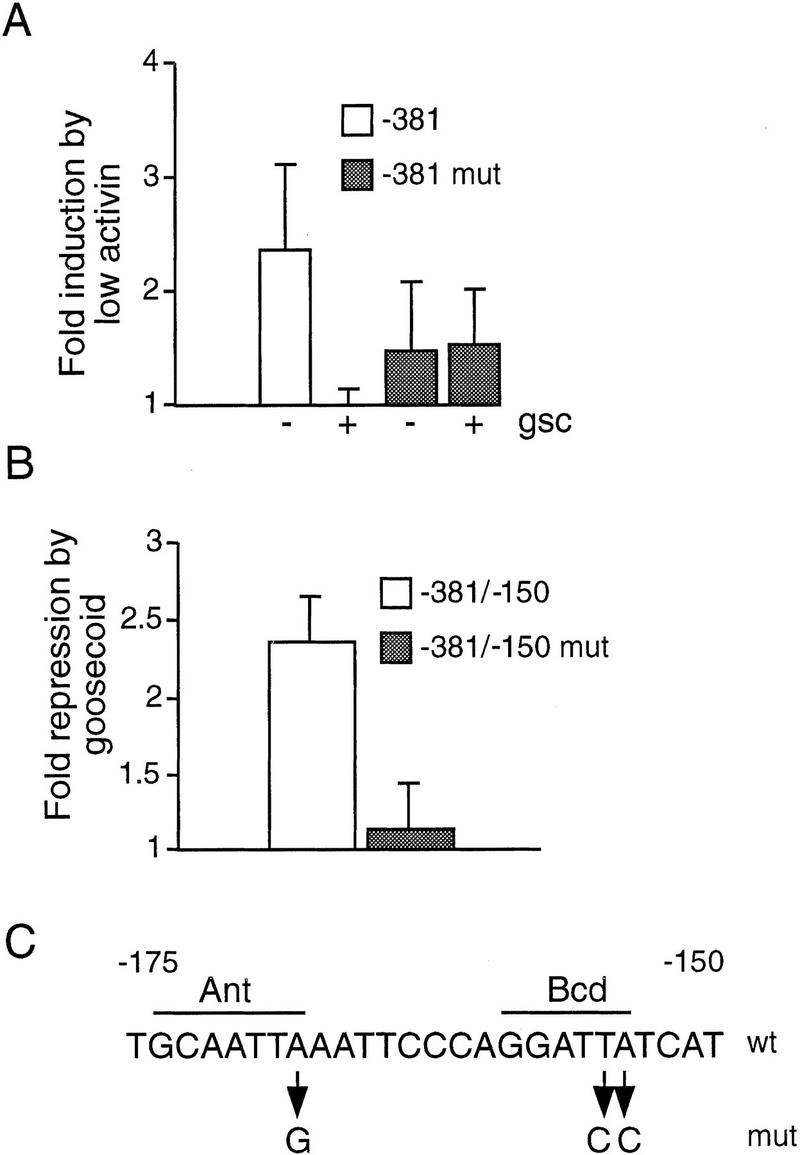
Suppression of −381Xbra2.luciferase activity by goosecoid requires Antennapedia and Bicoid homeodomain-binding sites. (A) Embryos were injected with 15 pg mutated −381Xbra2.firefly luciferase (see C), 15 pg −381Xbra2.Renilla luciferase, and 30 pg pEF-1α.β-galactosidase; in addition to this combination of DNAs, some embryos were injected with 1 pg of activin mRNA or with a mixture of 1 pg of activin mRNA and 1 ng of goosecoid mRNA. Animal caps were excised at the blastula stage and incubated until stage 12.5, when firefly and Renilla luciferase and β-galactosidase activities were determined. The graph shows fold induction by activin in the presence or absence of goosecoid, as obtained with the wild-type and mutated reporters. Normalized luciferase activities from four independent experiments were used in this figure. Bars show s.d.s. (B) NIH-3T3 cells were cotransfected in duplicate with a combination of CMV overexpression vector (either empty or goosecoid expressing), wild-type or mutated Xbra2/SV40 reporter, and reference plasmid. Luciferase activities were determined, and the results are expressed as the fold repression of reporter activity by goosecoid for each construct. Data from four experiments was used for this graph. Bars show s.d.s. (C) Sequence of the wild-type and the mutant Antennapedia and Bicoid sites.
