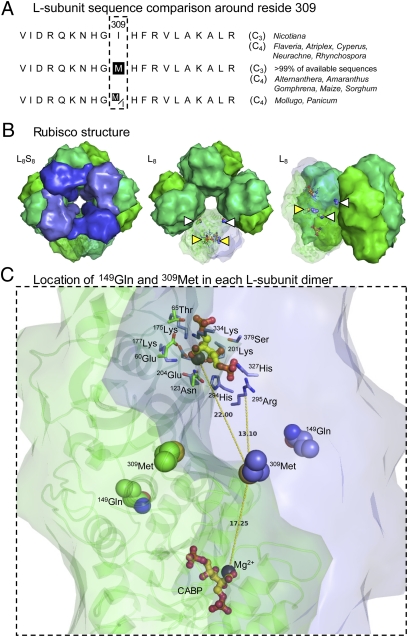
Fig. 4.
Conservation and location of the L-subunit residues 149 and 309 in higher plant rubisco. (A) Coding matrix summary of ClustalW-aligned residues 300–319 in L-subunit sequences from data sets with corresponding C3 and C4 speciation data (11, 20, 38). (B) Structure of spinach L8S8 rubisco and L8 core (L-subunits are shown in green; S-subunits are shown in blue) viewed from the top, showing central solvent channel (Left and Center), and from the side (Right). The relative locations of 149Gln (white triangle) and 309Met (yellow triangle) in one L-subunit pair (L2) is shown. The 309Met residues are located at the interface of L-subunits; the 149Gln residues are positioned at the L2–L2 interface toward the surface of the central solvent channel. (C) View of an L-subunit pair (L1 in green showing ribbon structural detail; L2 in blue) showing the positioning of 149Gln and 309Met relative to each Mg2+ (black sphere) and the reaction-intermediate analog 2-CABP (yellow and red ball and stick) bound to the two active sites in the dimer. The conserved active-site residues are shown for one active site. Distances (in Å) from the S atom of 309Met in L2 to each Mg2+ and the Cα atom of the closest conserved active-site residue, 295Arg, are shown. (This figure was prepared with PyMOL using the PDB co-ordinates 8RUC.)