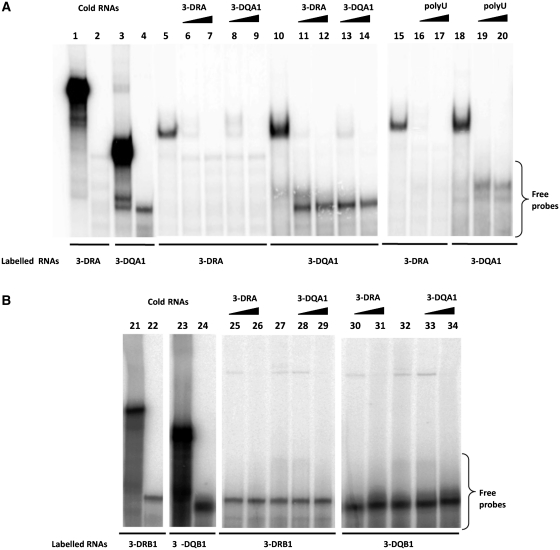
Figure 2.
3′UTR of MHC II mRNAs binding by REMSA. (A) REMSAs experiments performed using 3-DRA (lane 1) and 3-DQA1 (lane 3) riboprobes; lanes 2 and 4 show the digestion of riboprobes with T1 RNase. Lanes 5 and 15 show bands of interaction of M14 extract with 3-DRA, lanes 10 and 18 with 3-DQA1; competition experiments of 3-DRA binding were performed using cold 3-DRA (lanes 6 and 7), cold 3-DQA1 (lanes 8 and 9) and poly(U) homopolymers (lanes 16 and 17). Competition experiments of 3-DQA1 binding were performed using cold 3-DRA (lanes 11 and 12), cold 3-DQA1 (13 and 14) and poly(U) homopolymers (lanes 19 and 20). (B) REMSAs experiments carried out using 3-DRB1 (lane 21) and 3-DQB1 (lane 22) riboprobes. Lanes 22 and 24 show the digestion of riboprobes with T1 RNase; lanes 27 and 32 show bands of interaction of M14 extract with 3-DRB1 and 3-DQB1.Competition experiments of 3-DRB1 binding were performed using cold 3-DRA (lane 25 and 26) and cold 3-DQA1 (28 and 29). Competition experiments of 3-DQB1 binding were performed using cold 3-DRA (lanes 30 and 31) and cold 3-DQA1 (33 and 34).