Abstract
microRNAs (miRNAs) spatio-temporally modulate gene expression; however, very little is known about the regulation of their expression. Here, we hypothesized that the well-known cis-regulatory elements of gene expression, scaffold/matrix-attachment regions (MARs) could modulate miRNA expression. Accordingly, we found MARs to be enriched in the upstream regions of miRNA genes. To determine their role in cell type-specific expression of miRNAs, we examined four individual miRNAs (let-7b, miR-17, miR-93 and miR-221) and the miR-17–92 cluster, known to be overexpressed in neuroblastoma. Our results show that MARs indeed define the cell-specific expression of these miRNAs by tethering the chromatin to nuclear matrix. This is brought about by cell type-specific binding of HMG I/Y protein to MARs that then promotes the local acetylation of histones, serving as boundary elements for gene activation. The binding, chromatin tethering and gene activation by HMG I/Y was not observed in fibroblast control cells but were restricted to neuroblastoma cells. This study implies that the association of MAR binding proteins to MARs could dictate the tissue/context specific regulation of miRNA genes by serving as a boundary element signaling the transcriptional activation.
INTRODUCTION
The importance of nuclear architecture in governing transcription of genes/gene clusters in the genome is well established. This is achieved at multiple levels by (i) making distinct territories of chromosomes (1) and (ii) further compartmentalization of genes into distinct domains by periodic looping of chromatin, called as scaffold/matrix-attachment regions (S/MARs) (2,3). MARs tether the chromatin to the base of nuclear matrix, where transcription factories are present and poise the chromatin for transcription (4,5). Previous studies have demonstrated the importance of these cis-acting elements in viral integration, locus control of gene clusters in a spatio-temporal manner, promoting or inhibiting transcription of individual genes in a context-dependent manner (4,6–10). Regions of the genome attach to the nuclear scaffold/matrix in both a cell type- and cell cycle context-specific manner, although the precise mechanism(s) are yet unknown. It has been estimated that approximately 64 000 S/MARs divide the somatic genome into a series of ∼100 kb domains, with each domain bound by a S/MAR at each end. However, only a subset of potential S/MARs may be active in a cell at any given time. Changes in the activity of these sites may provide a means to modulate phenotype (11). This can be partly attributed to the binding of specific transcription factors/chromatin modifiers to these regions in the genome, thereby imparting gene regulatory function. It is, therefore, not surprising that changes in the nature of these MAR binding proteins (MARBPs) often lead to malignancies (12). Several studies have highlighted the importance of MARBPs like SATB1, SMAR1, NMP, SAF-A and other non-conserved AT hook region binding proteins like HMG family members in gene regulation and cancer (13–17). However, a categorical binding site representation has not been elucidated due to the non-conserved nature of these MAR sites. It is postulated that these MARBPs are crucial to the epigenetic regulatory circuits governed by cis-acting elements like S/MARs.
Since their discovery, microRNAs (miRNAs) have generated immense interest as modulators of gene expression in a tissue and organ-specific manner (18–21). A number of studies have been and are being pursued to understand their biogenesis and functions in regulating cell lineage commitment and disease manifestation. Since miRNAs complimentarily bind to their target mRNAs thereby interfering with translation and/or inducing degradation of target mRNAs, their activation can lead to inactivation of many target genes and hence pathways. Studies on miRNA-expression profiles of human tumors have identified signatures associated with diagnosis, staging, progression, prognosis and response to treatment (22–24). Recent reports also suggest complex interactions of miRNAs with the machinery that controls the transcriptome, concurrently targeting multiple mRNAs (24). Hence, understanding the regulation of expression of these miRNAs in normal and transformed tissues/cells is paramount, as this would provide fundamental insights into cancer cell reprogramming.
In this study, we hypothesized that cis-acting MAR elements would regulate the tissue and cell type-specific expression of miRNAs. We show that MARs are enriched in the upstream regions of miRNA genes (encoding primary transcript) and they spatially restrict the activation of miRNA genes as well as miRNA gene clusters. Further, using neuroblastoma cells and primary fibroblasts, we show that MARs can define the tissue/cell-specific expression of miRNAs augmenting or blocking their transcription. This regulation is governed by binding of a specific MAR binding protein HMG I/Y, a chromatin modulator.
MATERIALS AND METHODS
In silico prediction of MARs
MARs have been predicted to occur in every 10 kb in the mammalian genome (25,26). Thus, sequences spanning miRNA genes and a 10 kb upstream region were downloaded for all human miRNAs obtained from miRBase 14.0 and checked for the presence of conserved MAR (peak cut off = 0.5) using MARWiz (27). The conserved positions were marked and the window of the high MAR potential recorded.
Cell lines and transfections
The human neuroblastoma IMR-32 cells and mouse embryonic fibroblasts (MEFs) were cultured in RPMI and DMEM medium, respectively, supplemented with 10% FBS, l-glutamine and Pencillin–Streptomycin and maintained in a 5% CO2 humidified chamber at 37°C. Different concentrations of HMG I/Y siRNA were transfected using SureFECT transfection reagent (Qiagen) in IMR-32 cells for 72 h and evaluated for cell death, or processed for RT–PCR. Cell death was assessed by trypan blue exclusion assay.
PCR amplification and cloning
Regions surrounding the high MAR potential were amplified from IMR-32 genomic DNA using primers for the following miRNA-MARs:
let-7b F: AGATTTCCCTGCGTGTGAAG (Ta = 51)
let-7b R: AGGAGAGGCATTGACGAAGA
miR-221 F: GGGCAGGGTTTGTTTCTAGG (Ta = 51)
miR-221 R: TCAATGGAATTGCAACACAAA
miR-17 F (US1): GGGCACATTATACGTGCTTG (Ta = 51)
miR-17 R (US1): AAAACCTAGTCATGCCACCA
miR-93 F: TTCCAACAACTCTGCCATTTT (Ta = 51)
miR-93 R: TGTGCTGGGACAACTGGATA
miR-17–92 cluster US2 F: TGGCATTGGCTCTTTGATCAGCA(Ta = 56)
miR-17–92 cluster US2 R: TGCAAAAGTCCTGCATGGTTTGGT
All the experiments were performed using limiting cycles of PCR.
Matrix–loop partitioning assay
Nuclear matrix- and loop-associated pools of genomic DNA were prepared as previously described (28), with minor modifications. Briefly, IMR-32 and MEF cells were washed with phosphate buffer followed by sequential lysis with CSK-1 (0.5% Triton X-100, 10 mM PIPES at pH 6.8, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM PMSF and 1 x protease inhibitor cocktail) and CSK-2 buffers (same as CSK-1 except that Triton X-100 is omitted). DNase1was added and digestion was performed for 1 h. Digested supernatant (loop DNA), as well as pellet containing undigested material (nuclear matrix + MARs), were collected independently and DNA was purified by proteinase K digestion followed by phenol–chloroform extraction and ethanol precipitation. Isolated pools of matrix- or loop-associated DNA were used as templates for PCR amplification with different sets of primers designed for the miR-17–92 cluster and individual miRNAs. PCR products were resolved by agarose gel electrophoresis, stained with Ethidium bromide (Molecular Probes) and visualized by UV transillumination.
MAR binding assay
Nuclear matrix isolated from IMR-32 or MEF cells was suspended in 90 μl MAR binding buffer (20 mM Tris–HCl pH 7.4, 50 mM NaCl, 2 mM EDTA, 0.25 M Sucrose and 0.25 mg/ml BSA). Sheared salmon sperm DNA (100 μg/ml) and 5 ng/ml (50 000 cpm) biotin-labeled DNA fragments from MAR region of miRNAs were mixed and incubated with nuclear matrix fraction at 25°C for 4 h with constant gentle shaking. Reaction mixture was diluted with 1 ml binding buffer, centrifuged and the matrix-bound fragments were solubilized in 0.5% SDS. The soluble mixture was treated with 0.5 mg/ml proteinase K for 5 h, phenol–chloroform extracted, ethanol precipitated and finally resolved on an 8% polyacrylamide–0.1% SDS gel and documented with chemiluminescence assay kit (Pierce).
Chromatin immunoprecipitation assay
For the Chromatin immunoprecipitation (ChIP) assay, 1–5 × 106 cells were treated with DMEM containing 1% formaldehyde for 10 min at 37°C for cross-linking, which was stopped by a 10 min incubation with 1.5 M glycine. After washing twice, the cells were resuspended in 300 μl of SDS lysis buffer [50 mM Tris–HCl (pH 8.0), 10 mM EDTA, 1% SDS and protease inhibitors] by pipetting and kept on ice for 20 min. The chromatin was then sonicated into fragments with an average length of 0.3–7 kb. After centrifugation at 13 000 rpm for 10 min, the supernatants were diluted with dilution buffer [50 mM Tris–HCl (pH 8.0), 1.1% NP-40, 167 mM NaCl and protease inhibitors]. The extracts were pre-cleared by incubation with 30 μl of protein G-Sepharose beads (Amersham Biosciences) for 6 h. The supernatants were mixed with antibodies for 16 h and incubated with protein G-Sepharose beads for 3 h. The incubated beads were then washed once with low-salt buffer [50 mM Tris–HCl (pH 8.0), 2 mM EDTA, 2% NP-40 and 0.2% SDS] containing 150 mM NaCl, once with high-salt buffer containing 500 mM NaCl and once with LiCl wash solution [10 mM Tris–HCl (pH 8.0), 250 mM LiCl, 1 mM EDTA and 0.5% NP-40]. The washed beads were incubated in elution buffer [10 mM Tris–HCl (pH 8.0), 300 mM NaCl, 5 mM EDTA and 0.5% SDS] at 65°C for 12 h, followed by phenol–chloroform treatment and ethanol precipitation. ChIP DNA was amplified by standard PCR using Taq polymerase with the primers from Cybergene, as listed. The following antibodies were used: anti-HMG I/Y, anti-AcH3K9/14 (Santa Cruz), anti-H3K9 tri methyl (Abcam) and rabbit or mouse immunoglobins (Bio-Rad).
RESULTS
miRNA genes are flanked by MARs
The organization of chromatin into topologically constrained loops serves to define functional genetic domains. Such loops are thought to be established by S/MARs. MARs have been identified at the boundaries of functional transcription units from several species and they have been shown to buffer effects of flanking chromatin. However, whether MARs can buffer miRNA expression is unknown. A recent study showed that the upstream MARs serve as enhancers augmenting transcription while intra/intergenic MARs could function as transcription inhibiting structures (29).
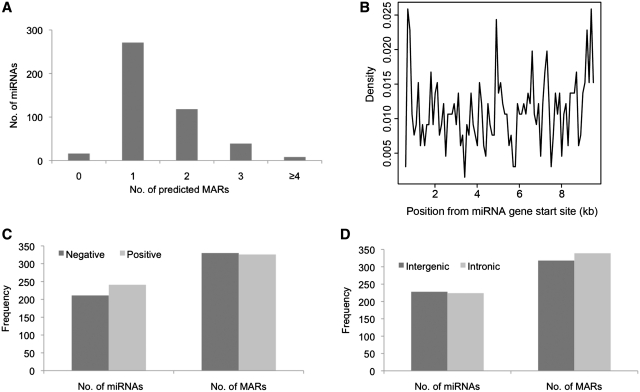
To identify if MARs were present upstream of and/or within miRNA genes, we examined the sequence spanning miRNA genes and a 10 kb region upstream using MARWiz. After filtering with high stringency parameters (peak threshold = 1), we found that 436 of 452 miRNAs considered here, possessed conserved upstream MAR elements, with ∼38% miRNAs possessing more than one MAR element (Figure 1A). MAR elements were found distributed across10 kb region upstream of miRNA genes, analyzed here. However, no MARs were detected within or encompassing the miRNA genes (Figure 1B). There were no significant differences in the presence of MAR elements among miRNAs coded by negative or positive DNA strands (Figure 1C) or in the intronic or intergenic miRNAs (Figure 1D). These observations imply that presence of an upstream MAR element is a conserved feature for miRNA coding genes. This led us to examine the functional consequence of the presence of these upstream MARs in regulating the miRNA expression.
Figure 1.
Properties of MARs predicted upstream of miRNAs. (A) Number of MAR elements predicted upstream of miRNAs. MARs were observed in the upstream regions of vast majority (96%) of miRNA genes. (B) Distance of predicted MARS from miRNA genes. MARs were predicted at distances as short as 610 bp to 9.5 kb upstream of the miRNA genes. (C) DNA strand specificity of miRNAs and predicted MARs. miRNAs on both positive and negative strands have similar frequency of upstream MARs. (D) Intronic and intergenic miRNA genes have similar frequency of upstream MARs.
The miRNA–MARs partition genes to active fronts
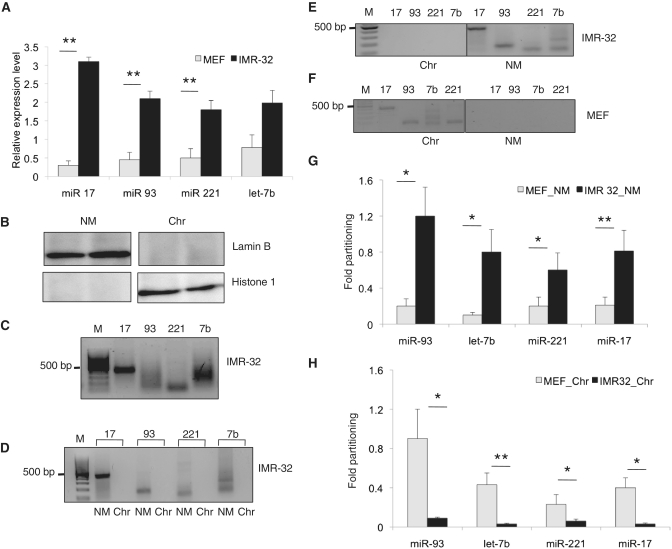
As MARs can regulate the tissue expressivity based on its tethering to nuclear matrix, we aimed to investigate the possible correlation between the cell type-specific expression of miRNAs to the presence of MARs. For this, we identified and characterized MARs from the miRNAs overexpressed in neuroblastoma and compared them to primary MEFs (Figure 2A). These include miR-221, let-7b, miR-17 and miR-93. All these miRNAs possessed MARs, however, at varying distances from the miRNA genes (Supplementary Figure S1). We then tested the functional activity of these MARs by checking their affinity to isolated nuclear matrix from IMR-32 cell line. For this, we fractionated the cells into nuclear matrix and chromatin fractions. Cross-contamination of each of the fractions was ruled out by use of Lamin B and histone 1 antibody (Figure 2B). MAR binding assays showed that all the MARs (amplified as single products from genomic DNA of IMR-32 cells, Figure 2C) bound to the nuclear matrix with equal propensity (Figure 2D). In vivo partitioning assays also revealed a strong association of these MARs with the nuclear matrix and not with the chromatin fraction in these cells (Figure 2E).
Figure 2.
MAR partitioning and expression of miRNAs. (A) Real-time PCR analysis showing the relative expression of indicated miRNAs in IMR-32 neuroblastoma cells and MEFs normalized to 5 s rRNA levels. The values are mean ± SD from three independent experiments. **Indicates P < 0.01. (B) Immunoblot analysis for nuclear matrix (NM)-specific lamin B and chromatin-specific histone 1 (H1) in each of the fractions to rule out cross contamination. (C) Amplification of the specific MAR element from each of the represented miRNAs from genomic DNA of IMR-32 cells. (D) MAR binding assay employing NM from IMR-32 cells and the biotin-labeled individual MAR elements as probe. PCR amplification of the individual MAR elements in the chromatin (Chr) or NM fractions of IMR-32 neuroblastoma cells (E) or mouse embryonic fibroblasts (MEFs) (F). The graphs in (G) and (H) depict the relative amount of MAR amplicons in NM and chromatin fractions of IMR-32 and MEF cells respectively, drawn of densitometric analysis from 3 independent experiments. Statistical comparisons were done using Students t-test. *Indicates P < 0.05 and **P < 0.01.
Since MARs are known to be cell type-specific and at a given point in any cell type only a subset of MARs bind to nuclear matrix and partition chromatin into distinct domains based on the genes to be transcribed, we checked if this partitioning was intact in MEFs. We observed that the MAR elements are amplified in the chromatin fraction and do not partition discretely into the nuclear matrix (Figure 2F). This is, in contrast to IMR-32 cells, where a distinct partitioning to nuclear matrix was observed. This was demonstrated by a relative partitioning analysis, where the amplicons were quantitated and compared in the nuclear matrix and chromatin fractions in IMR-32 and MEF cells, respectively (Figure 2G and H). By employing luciferase reporter assays, we found that the MARs upstream of miRNAs can augment the transcription, as has been previously observed for MARs upstream of protein coding genes (8,12) (Supplementary Figure S2). A luciferase vector hosting an extended region flanking the MARs excluding the core 200 bp MAR sequence served to negate random sequence activity, if any.
MARs as small range locus control regions for miRNA clusters
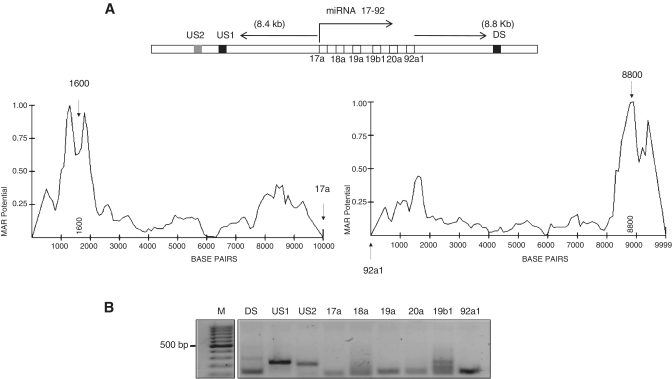
MARs are known to be insulators or enhancers for gene loci. In this case, we sought to check if a single MAR is sufficient to regulate the expression of all the miRNAs in a miRNA cluster. For this, we checked for the presence of MARs in the miR-17–92 cluster, containing 6 miRNAs namely miR-17, 18a, 19a, 20a, 19b1 and miR-92a1, all of which are expressed in neuroblastoma (Figure 3A). The 10 kb region flanking this miRNA cluster comprised of two conserved MARs-one upstream (depicted as US1) and one downstream (DS). Genomic PCR confirmed the amplification of the whole sequence cluster as individual segments (Figure 3B). We chose a non-MAR containing region 1 kb upstream of the US1 site (depicted as US2) and used this as control for elucidating MAR specific functions.
Figure 3.
MARs influence miRNA clusters. (A) Schematic representation of miRNA 17–92 cluster and the flanking 10 kb regions. The dark boxes represent conserved MAR elements (US1 and DS), boxes in white the individual miRNA genes and the shaded box a control region upstream of the MAR element (US2). The graph below depicts the MAR potential of the individual MAR element present upstream and downstream of the miRNA 17–92 cluster, with the arrows pointing the region of maximum peak potential. (B) Amplification of the MAR elements and the individual miRNA genes from the genomic DNA of IMR-32 cells.
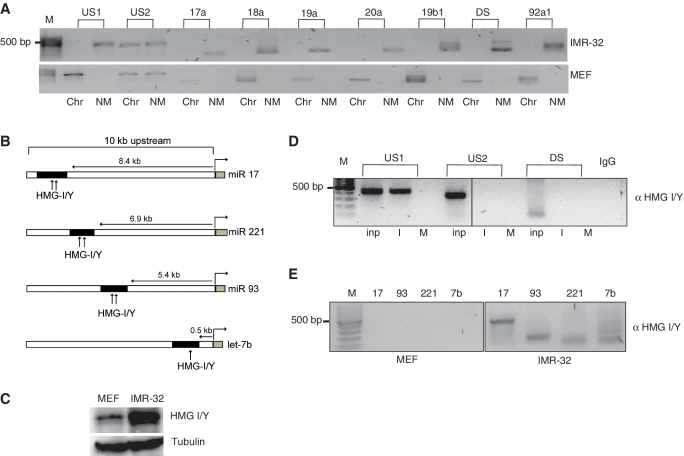
Next, we checked the partitioning of these miRNA genes into active matrix and chromatin loops and found that the MARs and the genes in the miR-17–92 cluster associated preferentially to nuclear matrix in IMR-32 cells, while in MEFs they were found to be associated with the chromatin fraction (Figure 4A).
Figure 4.
Factor binding and MAR partitioning. (A) NM and Chr from IMR-32 cells and MEFs were fractionated and checked for amplification of the individual genes and the MAR elements of the miRNA 17–92 cluster. (B) Schematic representation of the putative HMG I/Y binding sites (represented by arrows) in the MAR regions (black box) of the individual miRNA genes and the miRNA 17–92 cluster (gray boxes). The distance of MARs from the each miRNA gene is also provided. (C) Immunoblot analysis of HMG I/Y in 50 µg of total lysates from MEFs and IMR-32 cells. Tubulin was used as loading control. (D) Chromatin immunoprecipitate from IMR-32 (I) or MEFs (M) were analyzed for binding of HMG I/Y protein to the upstream or downstream MAR region from miRNA 17–92 cluster. Rabbit immunoglobin served as negative control (IgG). A quantity of 10% of the total lysate was used as input (Inp). (E) HMG I/Y binding to the individual MAR elements of miRNAs 17, 93, 221 and let-7b was checked in MEFs and IMR-32 cells.
The binding of MARBPs to MARs is critical for the regulatory role of these cis-acting elements, constituting a cis-element–trans-factor interplay. Therefore, we sought to identify the common factors that could potentially bind to all these MARs. For this, we performed binding site analysis of transcription factors in these MAR elements using ConSite (30). Interestingly, all the sequences showed atleast one high potential (≥80% cut-off) HMG I/Y protein binding site (Figure 4B). Also, HMG I/Y is an AT hook binding protein and is overexpressed upon malignant transformations (15). Immunoblot analysis showed that the expression of HMG I/Y protein in MEF cells was ∼2.4-fold lesser compared to IMR-32 cells (Figure 4C). This was reflected in our ChIP experiments where HMG I/Y specifically bound to the US1 MAR sequence in IMR-32 cells while the binding was negligible in MEF cells. Interestingly, the downstream MAR did not show any binding (Figure 4D). Rabbit immunoglobin and US2 served as negative controls. Similar to our observations for the miR-17–92 cluster MAR, we found that the individual MARs upstream of miRs let 7 b, 93 and 221 showed strong binding to HMG I/Y in IMR-32 cells and not in MEFs (Figure 4E).
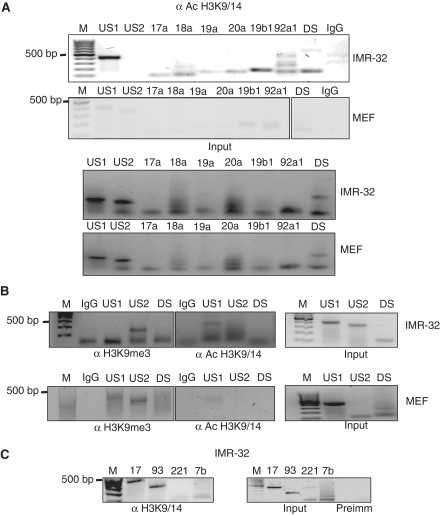
Since HMG I/Y proteins are minor groove AT hook binding proteins and are documented to be chromatin modulators, we checked the nature of chromatin in these sequences. In case of miR-17–92 cluster, all the individual miRNA genes were acetylated at histones H3 at lysine 9 and 14 positions (Figure 5A). The MAR sequences, both upstream and downstream, were also found to be acetylated at histone 3 in the same positions. US2 did not show any enrichment of acetylated histones, suggesting that hyperacetylated MARs may act as boundaries between chromatin in the expressed and non-expressed states (Figure 5A). However, in case of MEFs, the MARs were not enriched in acetylated histones, rather they were tri-methylated at histone lysine 9 position indicating a closed chromatin conformation (Figure 5B, lower panel). Individual MAR elements also displayed this effect where we could observe hyperacetylation marks in the MAR regions in IMR-32 cells (Figure 5C). Taken together, these results suggest the involvement of HMG I/Y in binding to the MARs of the miRNA genes and promoting an open chromatin conformation facilitating the miRNA gene transcription.
Figure 5.
MARs are active chromatin regions. (A) Chromatin from IMR-32 and MEFs were extracted and immunoprecipitated with acetylated histone 3 lysine 9/14. The immunoprecipitates were then analyzed for amplification of miR-17–92 cluster and the upstream and downstream MAR elements. Rabbit IgG served as negative control. Equal amounts of starting material were used, as verified using 10% total lysate as input. (B) Chromatin immunoprecipitates from IMR-32 and MEF cells were analyzed for histone 3 lys 9 trimethylation (H3K9 me3) or acetylation (H3K9/14 Ac) of the upstream and downstream MARs (US1 and DS) and control element (US2) to verify the epigenetic status of these regions. (C) Chromatin immunoprecipitate from IMR-32 cells were analyzed for the histone acetylation status of the MAR elements for the indicated miRNAs. A quantity of 10% of total lysate served as input. Pre-immune sera served as negative control.
HMG I/Y affects miRNA expression
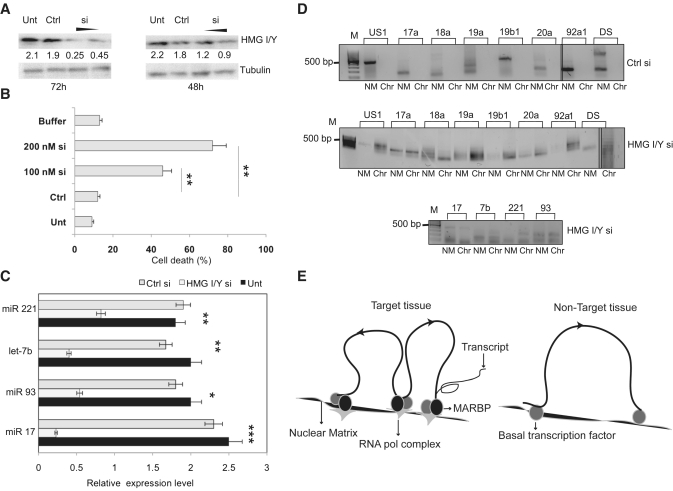
To further test, if the knockdown of HMG I/Y results in altered expression of the corresponding miRNA genes, we performed knockdown studies in IMR-32 cells. Interestingly, upon transfection of HMG I/Y siRNA in neuroblastoma cells most of the cells lost viability after 72 h of transfection. This time point coincides with 60–70% knockdown of HMG I/Y protein (Figure 6A and B). To address this, we opted for a time point when most of the cells were still viable but exhibited only a 30–40% knockdown. Under these conditions, the expression levels of individual miRNAs were quantified and we identified that HMG I/Y knockdown significantly reduced the expression of all the miRNAs, studied here (Figure 6C). Additionally, the partitioning of the MAR regions of these individual miRNAs at 48 h of knockdown was altered and we observed MARs both in the chromatin and nuclear matrix fractions even in limiting cycles of PCR (Figure 6D). Therefore, the increased cell death observed upon 60–70% knockdown might be a result of the down regulation of these anti apoptotic miRNA genes. These results demonstrate the crucial role of HMG I/Y in tethering the MARs to nuclear matrix, whereby miRNA gene expression is regulated.
Figure 6.
HMG I/Y regulates the miRNA gene partitioning. (A) Immunoblot analysis for HMG I/Y after partial knockdown using 100 and 200 nM siRNA, compared with siRNA control (Ctrl) and untreated cells, after 72 h and 48 h of transfection. Tubulin was used as loading control. (B) Quantification of cell death by trypan blue exclusion assay by transfection of HMG I/Y using 100 nM or 200 nM siRNA or Ctrl in IMR-32 cells after 72 h. Results are means of three independent experiments ± SD. (C) Quantitation of the levels of the indicated miRNAs in untreated cells IMR-32 cells (Unt) and after 48 h of siRNA mediated HMG I/Y knockdown (HMG I/Y si) using real-time RT–PCR analysis. The expression levels were normalized using 5s rRNA as internal control. A control siRNA (Ctrl si) was used as negative control. Statistical comparisons were done using Students t-test. *indicates P < 0.05, **P < 0.01, ***P < 0.001. (D) The NM and chromatin fractions from IMR-32 untreated control cells or HMG I/Y siRNA transfected cells were used to amplify the miRNA 17-92 gene cluster (panel above) and the MAR elements of indicated miRNAs (panel below). (E) Schematic representation of cis-regulation of miRNA expression by NM–loop organization in target and non-target tissues. In the target tissue (left panel) transcription is facilitated by attachment of MARs to the NM by specific MARBPs like HMG I/Y. This regulates expression by bringing the miRNA genes to the close proximity of transcription factories containing RNA polymerase and other components of the transcription apparatus. In non-target tissues/cells, MARs flanking the genes are not bound to the matrix due to absence of specific MARBPs preventing the gene/locus from associating with the focal points of high transcriptional activity.
DISCUSSION
This study presents evidence for miRNA gene regulation in a tissue/cell type-specific manner by cis-acting matrix-attachment regions. We have shown that the majority of the miRNA genes are flanked by at least one MAR element upstream of miRNA genes. This appears to be a conserved feature since the coding strand or the gene distribution with respect to protein coding genes (inter/intragenic) appears to have no bearing on the distribution of MARs upstream of miRNA genes. Further studies on MARs in promoters beyond 10 kb upstream of miRNA genes might provide additional insights into cis-regulatory roles of MARs. Also, we observed 96% of miRNA genes were predicted to contain MARs in the downstream region. This might represent potentiation of chromatin loop formation flanking the gene(s).
Models of DNA replication associated with the nuclear matrix have proposed that the replicon clusters correspond to the 50–200 kb repeating MARs that are attached to the nuclear matrix (31–34). As a basic unit, the loop is essential for DNA replication, transcription regulation and chromosomal packaging (35–37). Moreover, these MARs are dynamic and can vary from cell type to cell type, conferring another layer of spatio-temporal gene regulation. Here, we demonstrate the dependence of expression pattern of miRNA(s) and miRNA cluster on the association of upstream MARs with nuclear matrix. We used IMR-32 neuroblastoma cells as model to investigate some of the well-known oncomiRs including the miR-17–92 cluster. We show that the each of these miRNAs and the cluster possesses a MAR at different positions in the upstream regions (Supplementary Figure S1) and all these MAR elements can augment the transcriptional activity of these miRNA genes. Moreover, in neuroblastoma cells, these regions are strongly bound to the nuclear matrix. In primary fibroblasts, where the expression of the examined miRNAs was significantly reduced, MARs were associated with the chromatin. Also, the transcriptional activity of these MAR elements were significantly lesser in fibroblasts than in IMR-32 cells, suggesting the possible involvement of other factors in cell type-specific transcriptional augmentation.
It is widely believed that the biological significance and its behavior relies on determining the physical attributes of in vivo S/MARs and their association with specific proteins that bind to it. Though S/MARs function as the mediators of loop attachment, they are used in a selective and dynamic fashion. Also, while they are necessary they alone are not sufficient for chromatin loop formation (38). Nevertheless, their role in transcriptional regulation of genes and gene clusters is well appreciated. This transcriptional regulation is achieved by binding of epigenetic regulators that are ubiquitous and/or tissue-specific transcription factors including special (A+T)-rich binding protein 1 (SATB1), nuclear matrix protein 4 (NMP4) or CTCF (39). These proteins may promote the assembly of complex nucleoprotein supramolecular structures, as exemplified by the cage-like structures formed by SATB1, which circumscribe heterochromatic portions of chromosomes while allowing euchromatic sequences to loop out in the nucleus. MAR binding proteins may also recruit histone acetyltransferases and ATP-dependent chromatin remodeling complexes, thereby increasing gene expression (40). Thus, MAR elements may delimit chromosomal regulatory domains and recruit proteins that control the local chromatin structure.
Considering this option, we checked the putative transcription factor binding sites in all the individual MAR elements and identified HMG I/Y as the most likely MARBP. The HMG I/Y family of ‘high mobility group’ non-histone proteins consists of architectural transcription factors whose overexpression is highly correlated with both cancerous transformation and increased malignancy and metastatic potential of tumors in vivo. It is well documented that HMG I/Y regulates long-range, enhancer-dependent transcription on DNA and chromatin by changes in DNA topology (41).
We validated the HMG I/Y binding to the MAR sites of miRNA genes by use of ChIP. In case of miR-17–92 cluster, though the cluster was flanked on either side by MAR regions, HMG I/Y preferably bound to the upstream MAR. The binding of HMG I/Y was significantly less (almost negligible) in fibroblasts, which might be attributed to the lesser protein or cell type-specific post-translational modification. We believe that the cell type specificity might be conferred by proteins like HMG I/Y that recognize substrate structure, rather than nucleotide sequence and preferentially binds to distorted DNA structures. This would be then followed by a multiprotein assembly on DNA, forming a transcription enhancer code, defining cell type specificity. Since the MAR regions are generally enriched for transcription factor binding, it would be interesting to check the other conserved non MAR binding factor sites that lie in close proximity. For instance, we found that although Sox-5 and N-Myc are not classified as MAR binding proteins, their binding sites were juxtaposed to HMG I/Y binding sites in MAR elements.
In most instances, chromatin exerts a repressive effect and if either nucleosomes or inhibitory chromatin proteins (such as histone H1) are associated with important regulatory regions, gene transcription is attenuated or shut off. Several studies have shown that the binding to S/MARs by HMG-A promotes the establishment of an ‘open’ or accessible chromatin domain structure that is permissive for transcription to occur (42–44). Moreover these types of AT hook containing proteins play a central role in nucleosome remodeling complexes like RSC in Saccharomyces cerevisiae and SWI/SNF in humans (45,46). In this study, we found that the binding of HMG I/Y promotes an open chromatin conformation by hyperacetylating histones, favoring the transcription of miRNA genes. In fibroblasts we observed trimethylation at the histone 3 lysine 9 at the MAR loci, indicative of silenced chromatin. This was correlated to low expression levels of miRNA genes.
Since HMG I/Y can alter the topology of DNA we hypothesized that the knockdown of this protein by siRNA would affect the partitioning of MARs to the nuclear matrix fraction. With the near complete HMG I/Y knockdowns being lethal, we investigated partial knockdowns and found that even a 30–40% knockdown was sufficient to disrupt the discrete segregation of miRNA MARs to the nuclear matrix. Under these conditions, the expression levels of the miRNAs were also drastically reduced. Thus taken together these suggest that the binding of HMG I/Y to the MARs is crucial to their tethering to the nuclear matrix, which in turn influences the expression levels of miRNA(s).
Our studies for the first time have addressed the interplay between cis-acting MAR elements and miRNA gene expression as a function of cell/tissue type MAR binding proteins (Figure 6E). Therefore, the cell and context type specificity of miRNAs appear to involve a coordinated activity of cis-regulation, transcription factor binding, and chromatin modulation culminating in specific gene expression. Further understanding of this cell type-specific MARs and binding factors could provide valuable insights into the regulation of oncomirs and their networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: The Swedish Institute visiting researcher grant (to P.L.C.); the Swedish Research Council, Swedish Cancer Society; Swedish Childhood Cancer Research Foundation; BioCARE Region Skåne and Västra Götaland; ALF Funds at the VästraGötaland Region; the Assar Gabrielson Foundation, Sweden.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 2.Pienta KJ, Coffey DS. A structural analysis of the role of the nuclear matrix and DNA loops in the organization of the nucleus and chromosome. J. Cell Sci. Suppl. 1984;1:123–135. doi: 10.1242/jcs.1984.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- 3.Saitoh Y, Laemmli UK. Metaphase chromosome structure: bands arise from a differential folding path of the highly AT-rich scaffold. Cell. 1994;76:609–622. doi: 10.1016/0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 4.Bode J, Benham C, Knopp A, Mielke C. Transcriptional augmentation: modulation of gene expression by scaffold/matrix-attached regions (S/MAR elements) Crit. Rev. Eukaryot. Gene Expr. 2000;10:73–90. [PubMed] [Google Scholar]
- 5.Ottaviani D, Lever E, Mitter R, Jones T, Forshew T, Christova R, Tomazou EM, Rakyan VK, Krawetz SA, Platts AE, et al. Reconfiguration of genomic anchors upon transcriptional activation of the human major histocompatibility complex. Genome Res. 2008;18:1778–1786. doi: 10.1101/gr.082313.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martins RP, Ostermeier GC, Krawetz SA. Nuclear matrix interactions at the human protamine domain: a working model of potentiation. J. Biol. Chem. 2004;279:51862–51868. doi: 10.1074/jbc.M409415200. [DOI] [PubMed] [Google Scholar]
- 7.Goetze S, Baer A, Winkelmann S, Nehlsen K, Seibler J, Maass K, Bode J. Performance of genomic bordering elements at predefined genomic loci. Mol. Cell. Biol. 2005;25:2260–2272. doi: 10.1128/MCB.25.6.2260-2272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kulkarni A, Pavithra L, Rampalli S, Mogare D, Babu K, Shiekh G, Ghosh S, Chattopadhyay S. HIV-1 integration sites are flanked by potential MARs that alone can act as promoters. Biochem. Biophys. Res. Commun. 2004;322:672–677. doi: 10.1016/j.bbrc.2004.07.170. [DOI] [PubMed] [Google Scholar]
- 9.Farrell CM, West AG, Felsenfeld G. Conserved CTCF insulator elements flank the mouse and human beta-globin loci. Mol. Cell. Biol. 2002;22:3820–3831. doi: 10.1128/MCB.22.11.3820-3831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heng HH, Krawetz SA, Lu W, Bremer S, Liu G, Ye CJ. Re-defining the chromatin loop domain. Cytogenet. Cell Genet. 2001;93:155–161. doi: 10.1159/000056977. [DOI] [PubMed] [Google Scholar]
- 11.Kramer JA, Singh GB, Krawetz SA. Computer-assisted search for sites of nuclear matrix attachment. Genomics. 1996;33:305–308. doi: 10.1006/geno.1996.0198. [DOI] [PubMed] [Google Scholar]
- 12.Chattopadhyay S, Pavithra L. MARs and MARBPs: key modulators of gene regulation and disease manifestation. Subcell. Biochem. 2007;41:213–230. [PubMed] [Google Scholar]
- 13.Galande S, Kohwi-Shigematsu T. Linking chromatin architecture to cellular phenotype: BUR-binding proteins in cancer. J. Cell Biochem. Suppl. 2000;35(Suppl):36–45. [PubMed] [Google Scholar]
- 14.Malonia SK, Sinha S, Lakshminarasimhan P, Singh K, Jalota-Badhwar A, Rampalli S, Kaul-Ghanekar R, Chattopadhyay S. Gene regulation by SMAR1: Role in cellular homeostasis and cancer. Biochim. Biophys. Acta. 2011;1815:1–12. doi: 10.1016/j.bbcan.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Cerignoli F, Ambrosi C, Mellone M, Assimi I, di Marcotullio L, Gulino A, Giannini G. HMGA molecules in neuroblastic tumors. Ann. NY Acad. Sci. 2004;1028:122–132. doi: 10.1196/annals.1322.013. [DOI] [PubMed] [Google Scholar]
- 16.Fedele M, Fusco A. HMGA and cancer. Biochim. Biophys. Acta. 2010;1799:48–54. doi: 10.1016/j.bbagrm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Iorns E, Hnatyszyn HJ, Seo P, Clarke J, Ward T, Lippman M. The role of SATB1 in breast cancer pathogenesis. J. Natl Cancer Inst. 2010;102:1284–1296. doi: 10.1093/jnci/djq243. [DOI] [PubMed] [Google Scholar]
- 18.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. Chromosomal rearrangements and microRNAs: a new cancer link with clinical implications. J. Clin. Invest. 2007;117:2059–2066. doi: 10.1172/JCI32577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bode J, Goetze S, Heng H, Krawetz SA, Benham C. From DNA structure to gene expression: mediators of nuclear compartmentalization and dynamics. Chromosome Res. 2003;11:435–445. doi: 10.1023/a:1024918525818. [DOI] [PubMed] [Google Scholar]
- 26.Glazko GV, Koonin EV, Rogozin IB, Shabalina SA. A significant fraction of conserved noncoding DNA in human and mouse consists of predicted matrix attachment regions. Trends Genet. 2003;19:119–124. doi: 10.1016/S0168-9525(03)00016-7. [DOI] [PubMed] [Google Scholar]
- 27.Singh GB, Kramer JA, Krawetz SA. Mathematical model to predict regions of chromatin attachment to the nuclear matrix. Nucleic Acids Res. 1997;25:1419–1425. doi: 10.1093/nar/25.7.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Belle I, Cai S, Kohwi-Shigematsu T. The genomic sequences bound to special AT-rich sequence-binding protein 1 (SATB1) in vivo in Jurkat T cells are tightly associated with the nuclear matrix at the bases of the chromatin loops. J. Cell Biol. 1998;141:335–348. doi: 10.1083/jcb.141.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linnemann AK, Platts AE, Krawetz SA. Differential nuclear scaffold/matrix attachment marks expressed genes. Hum. Mol. Genet. 2009;18:645–654. doi: 10.1093/hmg/ddn394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandelin A, Wasserman WW, Lenhard B. ConSite: web-based prediction of regulatory elements using cross-species comparison. Nucleic Acids Res. 2004;32:W249–W252. doi: 10.1093/nar/gkh372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dijkwel PA, Mullenders LH, Wanka F. Analysis of the attachment of replicating DNA to a nuclear matrix in mammalian interphase nuclei. Nucleic Acids Res. 1979;6:219–230. doi: 10.1093/nar/6.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogelstein B, Pardoll DM, Coffey DS. Supercoiled loops and eucaryotic DNA replicaton. Cell. 1980;22:79–85. doi: 10.1016/0092-8674(80)90156-7. [DOI] [PubMed] [Google Scholar]
- 33.Berezney R. The nuclear matrix: a heuristic model for investigating genomic organization and function in the cell nucleus. J. Cell Biochem. 1991;47:109–123. doi: 10.1002/jcb.240470204. [DOI] [PubMed] [Google Scholar]
- 34.Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 35.Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int. Rev. Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- 36.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 37.Jackson DA. Chromatin domains and nuclear compartments: establishing sites of gene expression in eukaryotic nuclei. Mol. Biol. Rep. 1997;24:209–220. doi: 10.1023/a:1006873614521. [DOI] [PubMed] [Google Scholar]
- 38.Heng HH, Goetze S, Ye CJ, Liu G, Stevens JB, Bremer SW, Wykes SM, Bode J, Krawetz SA. Chromatin loops are selectively anchored using scaffold/matrix-attachment regions. J. Cell Sci. 2004;117:999–1008. doi: 10.1242/jcs.00976. [DOI] [PubMed] [Google Scholar]
- 39.Bidwell JP, Torrungruang K, Alvarez M, Rhodes SJ, Shah R, Jones DR, Charoonpatrapong K, Hock JM, Watt AJ. Involvement of the nuclear matrix in the control of skeletal genes: the NMP1 (YY1), NMP2 (Cbfa1), and NMP4 (Nmp4/CIZ) transcription factors. Crit. Rev. Eukaryot. Gene Expr. 2001;11:279–297. [PubMed] [Google Scholar]
- 40.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641–645. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 41.Bagga R, Michalowski S, Sabnis R, Griffith JD, Emerson BM. HMG I/Y regulates long-range enhancer-dependent transcription on DNA and chromatin by changes in DNA topology. Nucleic Acids Res. 2000;28:2541–2550. doi: 10.1093/nar/28.13.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao K, Kas E, Gonzalez E, Laemmli UK. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeves R. Structure and function of the HMGI(Y) family of architectural transcription factors. Environ. Health Perspect. 2000;108(Suppl. 5):803–809. doi: 10.1289/ehp.00108s5803. [DOI] [PubMed] [Google Scholar]
- 44.Reeves R, Leonard WJ, Nissen MS. Binding of HMG-I(Y) imparts architectural specificity to a positioned nucleosome on the promoter of the human interleukin-2 receptor alpha gene. Mol. Cell. Biol. 2000;20:4666–4679. doi: 10.1128/mcb.20.13.4666-4679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 46.Bourachot B, Yaniv M, Muchardt C. The activity of mammalian brm/SNF2alpha is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol. Cell. Biol. 1999;19:3931–3939. doi: 10.1128/mcb.19.6.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.