Abstract
Background
The prostate gland is the most common site of cancer and the second leading cause of cancer mortality in American men. It is well known that epigenetic alterations such as DNA methylation within the regulatory (promoter) regions of genes are associated with transcriptional silencing in cancer. Promoter hypermethylation of critical pathway genes could be potential biomarkers and therapeutic targets for prostate cancer.
Methods
This review discusses current information on methylated genes associated with prostate cancer development and progression.
Results
Over 30 genes have been investigated for promoter methylation in prostate cancer. These methylated genes are involved in critical pathways, such as DNA repair, metabolism, and invasion/metastasis. The role of hypermethylated genes in regulation of critical pathways in prostate cancer is reviewed.
Conclusions
These findings may provide new information of the pathogenesis of prostate cancer. Certain epigenetic alterations in prostate tumors are being translated into clinical practice for therapeutic use.
Introduction
Prostate cancer is the most common type of cancer (other than the skin) and the second leading cause of cancer mortality in American men. One man in 6 will develop prostate cancer during his lifetime, and 1 man in 34 will die of the disease.1 In 2010 in the United States, an estimated 217,730 new cases will be diagnosed, and 32,050 men will die of the disease.2 Although prostate cancer can be found early through PSA screening, this test is not 100% accurate, and false-positive results can lead to unnecessary prostate biopsy tests. However, a low mortality rate from prostate cancer suggests that public awareness of early detection and advanced treatments of prostate cancer have begun to affect prostate cancer outcomes.
The probability of developing prostate cancer sharply increases in the sixth decade of life (7%) and further increases after age 70 years (13%). These numbers contrast significantly with the probability of 0.01% among men under 40 years of age and 2.5% among those 40 to 59 years of age.2 The aging of the current population means that the disease will become an even greater public health problem in the future.
In some patients with prostate cancer, the disease progresses relatively slow. In these cases, patients often die with prostate cancer rather than of prostate cancer. However, some cases grow aggressively and metastasize through the bloodstream and the lymphatic system to other parts of the body. There are two important clinical challenges. The first challenge is the early detection of prostate cancer. Currently, digital rectal examination and serum prostate-specific antigen (PSA) screening are two main clinical diagnostic tools. However, due to their limited accuracies, these methods cannot reliably identify early-stage prostate cancer. Therefore, the identification of biomarkers that can facilitate the diagnosis of prostate cancer at the early stages could improve the current standard of treatments. The second challenge is to determine if a patient is presenting with aggressive or indolent prostate cancer. This is critically important information, given the significant morbidity associated with treatment interventions, and could eventually help distinguish men who need intensive treatment from those who may be better served by watchful waiting. Currently, the level of PSA, the clinical stage, and the grade of tumor (Gleason score) are used to estimate prognosis and determine treatment modalities. Although they are useful, additional biomarkers are needed to better predict the outcome of prostate cancer. Therefore, molecular biomarkers should help in determining who may need a prostate biopsy, which treatments a patient will undergo, and who may have a recurrence.
Role of DNA Methylation in Prostate Cancer
Tumorigenesis and progression of prostate cancer are results of the accumulation of genetic and epigenetic alterations. Although genetic changes are involved in the inactivation of genes with important anticancer functions (eg, tumor suppressor and DNA repair genes), DNA methylation in a promoter region is an important epigenetic mechanism for the downregulation (silencing) of expression of these genes. DNA methylation in the promoter region of tumor suppressor genes appears to occur at early stages of carcinogenesis and arises with various frequencies. Therefore, epigenetic changes have the potential to be a new generation of biomarkers. Several types of epigenetic changes have been reported for prostate cancer including DNA hypermethylation, loss of imprinting, and altered histone modification patterns.
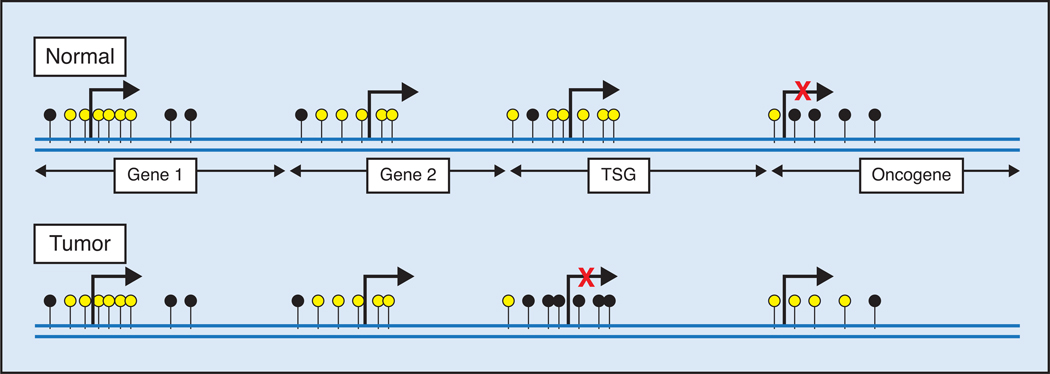
DNA methylation in the promoter of a number of genes occurs frequently in prostate tumors but rarely in normal prostate tissues. CpG islands are CpG-rich areas of 200 base-pairs to several kilo bases in length, usually located near the promoters of highly expressed genes, and they are the sites of almost all hypermethylation in human tumors,3 including the prostate. A common molecular feature associated with tumorigenesis is hypermethylation of cytosines 5′ to guanosines (CpG) within the regulatory (promoter) region of suppressor gene genomic DNA.4–8 5-Methyl cytosine is unstable and mutates to thymine, and methylated CpG sites degrade to TpG/CpA. In tumors, many CpG islands exhibit aberrant hypermethylation, resulting in gene silencing (Figure). Many of the silenced genes encode proteins that are tumor suppressor genes involved in tumorigenesis and progression.
Figure.
Role of DNA methylation in expression. Unmethylated and methylated CpG sites are indicated by yellow and black circles, respectively. Gene 1 and gene 2 are rarely methylated and therefore expressed. Densely hypermethylated CpG islands in the promoter region of a tumor suppressor gene (TSG) in tumor inhibit expression. Hypomethylation in the promoter region of oncogene in tumor reactivates a transcription process.
DNA Methylation Detection Methods
Multiple molecular biology techniques are available for detecting the DNA methylation pattern in genomic DNA.9 Based on the type of technique used, two major groups of detection methods are available.
Hybridization Method
This method is a combination of Southern blot and methylation-sensitive restriction enzymes treatment together with polymerase chain reaction (PCR). Since some restriction enzymes are methylation-sensitive, these enzymes cannot digest methylated target sequence. Combined with Southern blot technique, the hybridization method can assess the overall methylation status of target CpG sites. Major limitations of this technique are requirement of a large amount of genomic DNA and limited information of promoter methylation. At least 5 µg of genomic DNA is needed to analyze the methylation status. Results from this technique provide information for target CpG sites of methylation-sensitive restriction enzymes.9
PCR Method
In order to detect methylated CpG sites, DNA samples are modified by sodium bisulfite. Sodium bisulfite deaminates cytosine and transforms into uracil.10 Methylated cytosine, however, is not transformed by bisulfite treatment. Currently, methylation-specific PCR (MSP) and quantitative real-time MSP are two major techniques detecting methylation with the use of bisulfite-modified DNA. These PCR methods can be performed in a short time without a large amount of DNA sample. Also, they provide specific and sensitive results, especially using quantitative real-time MSP, which has sufficient sensitivity to detect methylation of 0.1% of alleles. However, like other PCR-based techniques, these methods may provide false-positive results.11
Hypermethylated Genes in Prostate Tumor
The majority of previous publications in epigenetic research in prostate cancer focused on DNA hypermethylation. Indeed, gene-silencing is more common by DNA hypermethylation in the promoter region than by DNA mutations in carcinogenesis. Numerous studies on various hypermethylated genes in different cancers suggest a key part of the carcinogenesis and progression of cancer.12
Currently, over 30 genes have been investigated for their frequencies of hypermethylation and for their potential role in prostate cancer (Table). Many of these hypermethylated genes are tumor suppressor genes that are coded for the proteins that regulate the cell cycle and/or promote apoptosis. The functions of tumor suppressor genes in prostate cancer fall into five major categories: DNA repair, apoptosis, cell cycle, corticosteroid hormonal response, and invasion/metastasis. Defected function of these genes by promoter hypermethylation can contribute to the carcinogenesis and progression of prostate cancer.
Table.
Frequencies of Hypermethylated Genes in Prostate Cancer
| Gene | Common Name | Frequency (Methylated/N) | Reference |
|---|---|---|---|
| APC | Adenomatous polyposis coli | 27% (27/101) | Maruyama et al17 |
| 90% (66/73) | Yegnasubramanian et al19 | ||
| 57% (21/37) | Kang et al22 | ||
| 100% (118/118) | Jerónimo et al20 | ||
| 78% (88/113) | Florl et al28 | ||
| 82% (59/72) | Tokumaro et al116 | ||
| 64% (109/170) | Enokida et al117 | ||
| 3.0* | Rosenbaum et al48 | ||
| 48% (25/52)** | Hoque et al27 | ||
| 83% (44/53) | Bastian et al45 | ||
| 73% (131/179) | Cho et al66 | ||
| 27% (21/79) | Henrique et al115 | ||
| 83% (65/78) | Bastian et al55 | ||
| 40% (182/459) | Richiardi et al118 | ||
| AR | Androgen receptor | 13% (2/15) | Kinoshita et al78 |
| 25% (6/24) | Nakayama et al80 | ||
| 8% (3/38) | Sasaki et al79 | ||
| 15% (16/109) | Yamanaka et al18 | ||
| 39% (30/76) | Reibenwein et al81 | ||
| CAV1 | Caveolin-1 | 91% (20/22) | Cui et al130 |
| CCND2 | Cyclin D2 | 32% (32/101) | Padar et al42 |
| 99% (117/118) | Henrique et al43 | ||
| CD44 | CD44 antigen | 78% (31/40) | Lou et al56 |
| 68% (27/40) | Kito et al46 | ||
| 32% (36/111) | Woodson et al41 | ||
| 72% (58/81) | Singal et al39 | ||
| CDH1 | E-cadherin | 54% (19/35) | Li et al131 |
| 27% (27/101) | Maruyama et al17 | ||
| 0% (0/111) | Woodson et al41 | ||
| 69% (70/101) | Padar et al42 | ||
| 24% (22/90) | Woodson et al40 | ||
| 0% (0/73) | Yegnasubramanian et al19 | ||
| 4% (5/114) | Florl et al28 | ||
| 61% (49/81) | Singal et al39 | ||
| 77% (40/52)** | Hoque et al27 | ||
| 30% (6/20) | Yao et al91 | ||
| CDH13 | H-cadherin | 31% (31/101) | Maruyama et al17 |
| 45% (68/151) | Alumkal et al132 | ||
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (p16) | 13% (3/24) | Jarrard et al25 |
| 70% (21/30) | Gu et al26 | ||
| 73% (8/11) | Nguyen et al24 | ||
| 3% (3/101) | Maruyama et al17 | ||
| 66% (21/32) | Konishi et al21 | ||
| 6% (4/73) | Yegnasubramanian et al19 | ||
| 77% (91/118) | Jerónimo et al20 | ||
| 4% (5/113) | Florl et al28 | ||
| 37% (19/52) | Hoque et al27 | ||
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (p14) | 22% (2/9) | Nguyen et al24 |
| 3% (1/32) | Konishi et al21 | ||
| 6% (1/16) | Konishi et al34 | ||
| 4% (5/118) | Jerónimo et al20 | ||
| 0% (0/73) | Yegnasubramanian et al19 | ||
| 37% (19/52)* | Hoque et al27 | ||
| 6% (6/95)* | Rouprêt et al35 | ||
| DAPK | Death-associated protein kinase | 1% (1/101) | Maruyama et al17 |
| 36% (39/109) | Yamanaka et al18 | ||
| 0% (0/73) | Yegnasubramanian et al19 | ||
| 28% (27/95)** | Rouprêt et al35 | ||
| EDNRB | Endothelin receptor type B | 70% (23/35) | Nelson et al57 |
| 83% (40/48) | Jerónimo et al58 | ||
| 49% (36/73) | Yegnasubramanian et al19 | ||
| 72% (58/81) | Singal et al39 | ||
| 100% (80/80) | Ellinger et al59 | ||
| 50% (9/18)** | Bastian et al113 | ||
| EPHA7 | EPH receptor A7 | 42% (20/48) | Guan et al135 |
| ER-α | Estrogen receptor alpha | 90% (28/31) | Li et al89 |
| 19% (14/73) | Yegnasubramanian et al19 | ||
| ER-β | Estrogen receptor beta | 83% (19/23) | Nojima et al90 |
| 65% (13/20) | Yao et al91 | ||
| FHIT | Fragile histidine triad | 15% (15/101) | Maruyama et al17 |
| GSTP1 | Glutathione S-transferase P1 | 100% (20/20) | Lee et al96 |
| 91% (52/57) | Lee et al99 | ||
| 75% (24/32) | Santourlidis et al100 | ||
| 94% (16/17) | Goessl et al101 | ||
| 44% (4/9)** | Suh et al107 | ||
| 72% (23/32)** | Goessl et al106 | ||
| 91% (63/69) | Jerónimo et al102 | ||
| 79% (22/28) | Cairns et al98 | ||
| 85% (89/105) | Jerónimo et al104 | ||
| 36% (36/101) | Maruyama et al17 | ||
| 75% (24/32) | Konishi et al21 | ||
| 58% (7/12) | Gonzalgo et al103 | ||
| 71% (43/61) | Harden et al97 | ||
| 88% (96/109) | Yamanaka et al18 | ||
| 84% (99/118) | Woodson et al41 | ||
| 100% (18/18) | Köllermann et al105 | ||
| 95% (69/73) | Yegnasubramanian et al19 | ||
| 87% (32/37) | Kang et al22 | ||
| 95% (112/118) | Jerónimo et al20 | ||
| 72% (58/81) | Singal et al39 | ||
| 79% (89/113) | Florl et al28 | ||
| 48% (25/52)** | Hoque et al27 | ||
| 83% (79/95)** | Rouprêt et al35 | ||
| HIC1 | Hypermethylated in cancer 1 | 99% (108/109) | Yamanaka et al18 |
| 100% (73/73) | Yegnasubramanian et al19 | ||
| 89% (N/A) | Kekeeva et al60 | ||
| LPL | Lipoprotein lipase | 38% (21/56) | Kim et al49 |
| MGMT | O6-methylguanine DNA methyltransferase | 25% (8/32) | Konishi et al21 |
| 0% (0/101) | Maruyama et al17 | ||
| 2% (2/109) | Yamanaka et al18 | ||
| 19% (22/118) | Jerónimo et al20 | ||
| 76% (28/37) | Kang et al22 | ||
| 1% (1/73) | Yegnasubramanian et al19 | ||
| 19% (10/52)** | Hoque et al27 | ||
| 15% (14/95)** | Rouprêt et al35 | ||
| PITX2 | Paired-like homeodomain 2 | 3.4* | Weiss et al52 |
| NA | Vanaja et al54 | ||
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 88% (64/73) | Yegnasubramanian et al19 |
| 71% (38/53) | Bastian et al45 | ||
| 65% (51/78) | Bastian et al55 | ||
| 68% (54/80) | Ellinger et al59 | ||
| RARβ | Retinoic acid receptor, beta | 79% (11/14) | Nakayama et al93 |
| 53% (54/101) | Maruyama et al17 | ||
| 78% (85/109) | Yamanaka et al18 | ||
| 84% (42/50) | Zhang et al94 | ||
| 70% (79/113) | Florl et al28 | ||
| 40% (32/81) | Singal et al39 | ||
| 35% (18/52)** | Hoque et al27 | ||
| RASSF1A | Ras association domain family 1 isoform A | 71% (37/52) | Liu et al37 |
| 53% (54/101) | Maruyama et al17 | ||
| 99% (117/118) | Jerónimo et al20 | ||
| 49% (40/81) | Singal et al39 | ||
| 78% (88/113) | Florl et al28 | ||
| 96% (70/73) | Yegnasubramanian et al19 | ||
| 84% (31/37) | Kang et al22 | ||
| 73% (38/52)** | Hoque et al27 | ||
| 74% (97/131) | Kawamoto et al38 | ||
| RBP1 | Retinol-binding protein 1 | 81% (96/118) | Jerónimo et al20 |
| 47% (17/36) | Jerónimo et al109 | ||
| S100A2 | S100 calcium-binding protein A2 | 99% (117/118) | Jerónimo et al20 |
| 94% (32/34) | Rehman et al134 | ||
| SLC5A8 | Solute carrier family 5, member 8 | 70% (7/10) | Park et al11 |
| SLC18A2 | Vesicular monoamine transporter 2 | 88% (15/17) | Sørensen et al63 |
| TIG1 | Tazarotene-induced gene 1 | 55% (17/31) | Tokumaro et al110 |
| 53% (26/50) | Zhang et al94 | ||
| 70% (43/61) | Topaloglu et al111 | ||
| 70% (125/179) | Cho et al66 | ||
| 10% (16/168)† | Ellinger et al112 | ||
| 96% (77/80) | Ellinger et al59 | ||
| TIMP-2 | Tissue inhibitor of metalloproteinase-2 | 60% (25/42) | Pulukuri et al123 |
| TIMP-3 | Tissue inhibitor of metalloproteinase-3 | 6% (7/109) | Yamanaka et al18 |
| 97% (114/118) | Jerónimo et al20 | ||
| 0% (0/73) | Yegnasubramanian et al19 | ||
| 37% (19/52)** | Hoque et al27 | ||
| 41% (37/91)** | Rouprêt et al35 | ||
| TNFRSF10C | TNF receptor superfamily, member 10c | 50% (25/50) | Shivapurkar et al64 |
| 65% (117/180) | Cho et al66 | ||
| 78% (46/59) | Cheng et al67 | ||
Hazard ratio
Urine samples
Serum DNA
N/A = not available
DNA Repair Gene
Although the specific causes of prostate cancer are not known, androgens, estrogens, inflammation, and DNA repair capacity have been implicated. DNA is constantly damaged by endogenous oxygen free radicals and exogenous chemicals. DNA mutations are estimated to spontaneously occur 20,000 to 40,000 times every day.13,14 The DNA repair process is important to the survival of the cell. Therefore, different repair pathways are available to reverse the different types of DNA damage. More than 150 DNA repair enzymes participate in this process.15 Defects in these DNA repair pathways may increase persistent mutations in daughter cell generations, genomic instability, and ultimately a prostate cancer risk.
Among several distinct DNA repair pathways, the direct reversal repair pathway may be important in carcinogenesis in the prostate. Methylguanine DNA methyltransferase (MGMT), the only known enzyme in the direct reversal repair pathway, leads to the direct restoration of the natural chemical composition of DNA without the need for genomic reconstruction. Therefore, defective MGMT activity is associated with an increased mutation.16 Reports regarding MGMT methylation in prostate tumor tissues have been inconsistent. While three studies reported a low frequency of MGMT promoter hypermethylation (0% to 2%) in prostate tumor tissues,17–19 others observed higher prevalence of hypermethylation (19% to 76%).20–22 Two investigator groups reported 15% and 19% MGMT hypermethylation frequencies in urine sediment samples collected from prostate cancer patients.27,35 These data suggest that MGMT promoter methylation can be a potential biomarker for early detection and surveillance of prostate cancer. However, larger studies will be necessary to resolve these inconsistent results.
Cell Cycle Genes
The cell cycle pathway regulates cell growth. One of the distinguishing characteristics of tumor cells is uncontrolled growth. Many genes act as checkpoints that regulate the cell cycle. Defective cell cycle genes may lead to the carcinogenesis and progression of prostate cancer.23
The tumor suppressor gene CDKN2 is one of the cyclin-dependent kinase inhibitors (CDKIs). CDKN2A (p16INK4), a key protein in the signaling pathway, can be damaged by a variety of genetic and epigenetic changes including hypermethylation in prostate cancer. Aberrant CDKI expression is observed in many tumor tissues including prostate.20,21,24 Results regarding the frequency of CDKN2A promoter methylation are inconsistent in prostate tumors, ranging from 3% to 77%.17,19–21,24–28 Perhaps these inconsistent results are due to different detection methods and/or different targets of methylated loci. Since Herman et al29 first reported inactivation of CDKN2A by DNA methylation in prostate cancer, other researchers have investigated the role of hypermethylated CDKN2A in the carcinogenesis and progression of prostate cancer.17,19–21,24–28 Although there was no significant association between CDKN2A low expression and increased CDKN2A exon 2 methylation, the exon 2 methylation may be a potential biomarker for prostate tumor.24 These results were confirmed by other investigators. Konishi et al21 observed that methylation occurred in the promoter region in 9% of samples and in exon 2 in 66% of tumors. Jerónimo et al20 found that the CDKN2A gene was frequently methylated in tumor tissue (77%) and in benign prostatic hyperplasia (BPH). These data support p16 methylation as a potential biomarker for an early detection of prostate cancer.
Another CDKI, the CDKN2A (p14ARF) promoter, has been methylated in various cancers,30–33 including prostate cancer.19–21,24,27,34,35 Based on seven publications, frequencies of p14ARF methylation ranges from 0% to 37%.19–21,24,27,34,35 Without two outliers,24,27 most published studies reported low methylation rates that ranged from 0% to 6%.19,21,24,34,35 Thus, the p14 is not a good candidate for a biomarker.
The RAS family of proto-oncogenes plays a key role in signal transduction pathways involved in cellular proliferation and survival, interacting with other regulatory circuits of cell growth and death. RAS association domain family protein 1 isoform A (RASSF1A) is known as a tumor suppressor gene. The RASSF1 protein was known to be associated with the DNA repair proteins and with the apoptotic effect.36 Inactivation by methylation of RASSF1A may deregulate the DNA repair pathway and cell cycle control in the tumor. The RASSF1A gene is silenced by aberrant methylation of the promoter in a large fraction of various cancers including prostate.37 In prostate tumors, RASSF1A promoter methylation is a common event, occurring in 49% to 99% of tumor tissues.17,19,20,22,27,28,37–39 RASSF1A promoter methylation is also associated with aggressive prostate cancer.17,22,37
Others cell cycle genes — CD44, cyclin D2 (CCND2), lipoprotein lipase (LPL), endothelin B receptor (EDNRB), hypermethylated in cancer 1 (HIC1), paired-like homeodomain transcription factor 2 (PITX2), and prostaglandin-endoperoxidase synthase 2 (PTGS2) — are often have a lower expression in prostate tumor tissues than in adjacent normal tissues. These low expressions are significantly correlated with promoter methylation level.40–45 Furthermore, expression of these genes and their promoter methylation may correlate with the tumorigenesis, progression, and clinicopathological features of prostate cancer.46–55 Many studies observed relatively high frequencies of promoter methylation in these genes: CD44 (32% to 78%),39,41,46,56 cyclin D2 (32% to 99%),42,43 LPL (38%),49 EDNRB (49% to 100%),19,39,57–59 HIC1, (89% to 100%),18,19,60 PITX2,52,54 and PTGS2 (65% to 88%).19,45,55,59 The frequencies of methylation of these genes, with the exception of EDNRB and HIC1, were significantly higher in prostate tumors than in normal tissues.42,43,49,55,57 Together, promoter methylation of these genes is a good candidate as a useful prostate cancer biomarker for the identification of the more aggressive prostate cancer that might benefit from different therapeutic modalities. However, the methylation status of EDNRB and HIC1 in prostate tumors parallels the respective normal tissue, although a high proportion of tumors are methylated.18,19,39,58–60 Therefore, DNA methylation sites in EDNRB and HIC1 are not good candidates for a marker for prognostic marker for prostate cancer progression and an intervention target for prostate cancer.
Apoptosis Genes
Programmed cell death (apoptosis) is a critical process for carcinogenesis in human. Typical morphological characteristics of apoptosis are damages of the plasma membrane, condensation and fragmentation of the nucleus, and DNA fragmentation.61 A major component of the apoptosis pathway is the caspase family. However, other genes, including death-associated protein kinase (DAPK), fragile histidine triad (FHIT), solute carrier family 5A8 (SLC5A8), vesicular monoamine transporter 2 (SLC18A2), and tumor necrosis factor receptor superfamily, member 10C (TNFRSF10C), are also involved in this pathway. A repressed expression of these genes by hypermethylation in the promoter region has been shown for prostate cancer.17–19,35,62–65 However, DAPK and FHIT may have a limited value due to a persistently low frequency of methylation in tumors and normal tissues.17–19,35 SLC5A8, SLC18A2, and TNFRSF10C were found to be hypermethylated in 50% to 88% of prostate cancers and significantly downregulated in tumor compared with normal prostate tissues.11,62–64,66,67 Expression of SLC18A2 and TNFRSF10C is negatively associated with biochemical recurrence after radical prostatectomy.63,68
Corticosteroid Hormonal Response Genes
The specific causes of prostate cancer are not known, but multiple etiological factors, including genetics, hormones, diet, infection, and environmental exposures, are thought to play significant roles. Although the precise role of androgens and their receptors in the carcinogenesis and progression of prostate cancer has not been fully investigated, previous studies suggest that these genes are important.69,70 Differences in the activities of these enzymes are determined to a large extent by genetic and epigenetic changes in the genes encoding them.
It is known that androgens stimulate the growth of prostate cells through the androgen receptor (AR).71 While silencing of AR expression decreases growth and induces apoptosis in vitro,72–74 overexpression of the AR also induces growth inhibition and apoptosis.75 In addition to prostatectomy and radiation therapy, androgen deprivation is one of the most effective treatments for prostate cancer. However, many advanced prostate cancer cells can survive in a low androgen environment due to a high expression of the androgen receptor.76 AR is one of the most frequently overexpressed proteins in the androgen-independent cases.77 Feldman and Feldman76 suggested five different possible pathways that lead to development of androgen-independent status. Several groups found AR promoter methylation in 8% to 39% of the prostate tumor tissues.18,78–81 Frequencies of AR promoter methylation are higher in androgen-independent cases than in primary prostate tumor tissues.78,80
The role of estrogen in the carcinogenesis of prostate tissues is not clear. However, a loss of expression of the estrogen receptor (ER)-β was induced by promoter methylation during the development of prostate cancer.82 The biological actions of estrogens are meditated by the ER. Two ERs are highly homologous DNA-binding domains but different N-terminus and ligand-binding domains. Both ERs, ER-α and ER-β, are downregulated in prostate tumor tissues.83,84 Promoter methylation is the primary mechanism responsible for low expression of ERs.79,85,86 ER-α expression is associated with a poor prognosis for hormonal therapy.87 ER-β is the main subtype in the prostate tissue and may serve as a tumor suppressor gene since ER-β protects against uncontrolled cell proliferation in normal prostate cells.86 However, high expression of ER-β in prostate tumors is associated with increased risk for recurrence and distant metastasis.84,88 Therefore, ER-β may have multiple roles in carcinogenesis and progression. The frequency of ER promoter methylation ranges from 19% to 90% in prostate tumors.19,89–91 The extent of ER promoter methylation is significantly higher in prostate tumors than in the BPH samples.89,90
Retinoic acid receptor β (RARβ) is known as a tumor suppressor gene by interacting with retinoic acid. Expression of RARβ is reported to be absent or downregulated in tumor tissues,92 and the RARβ2 promoter is aberrantly methylated in many cancers, including prostate cancer.93 Several groups reported that frequencies of methylation of the RARβ2 promoter range from 40% to 84% of primary prostate cancers but rarely in normal prostate tissues or BPH samples.17,18,28,39,93,94 Moderately high frequency of RARβ promoter methylation was observed in 35% of urine samples.27 In addition, the RARβ2 promoter is methylated in 20% of prostatic intraepithelial neoplasia (PIN) samples. Therefore, RARβ2 gene methylation may be an ideal biomarker candidate for early detection of prostate cancer.18,93
Glutathione S-transferase P1 (GSTP1) is involved in the detoxifying process and elimination of potentially genotoxic foreign compounds by conjugating glutathione into toxic chemicals. These processes protect prostate cells from DNA adducts and carcinogenesis. Thus, defective GSTP1 activity may increase DNA mutations, thereby possibly increasing the risk of prostate cancer.95 Because of its consistently frequent hypermethylation in the promoter region in prostate cancer, GSTP1 is perhaps the most studied gene in prostate cancer. Lee et al96 first reported a high frequency of GSTP1 hypermethylation in prostate tumor tissues.96 Since then, numerous studies confirmed similar results. Methylation of the GSTP1 promoter region occurs in 36% to 100% of tumor tissues.17–22,28,39,41,96–105 However, this methylation is rarely detected in normal prostate or BPH tissues. GSTP1 methylation was also detected consistently with high frequency in urine samples, blood, and ejaculates of prostate cancer patients, while either low or no methylation was detected in the samples from healthy controls.27,25,106,107 These different frequencies of GSTP1 promoter hypermethylation between tumor and normal prostate tissues make an ideal biomarker for prostate cancer.
Retinoids have an antitumorigenesis function and are involved in cell growth and differentiation. Their functional effects are mainly mediated by retinol-binding protein (RBP1). The role of RBP1 expression in carcinogenesis is not yet defined. However, the low expression of RBP1 by promoter methylation has been associated with the malignant tumor tissues, including prostate.108,109 Two studies reported that RBP1 promoter hypermethylation was found in 47% and 81% of tumors. No BPHs and normal prostate tissues were methylated.20,109
Tazarotene-induced gene 1 (TIG1) is frequently silenced in prostate tumors. This gene, also known as retinoid acid (RA) receptor-responsive 1 gene, was first identified as an RA-responsive gene. Several investigators reported that TIG1 was frequently methylated (53% to 96%) in prostate tumors, but in normal tissue or benign hyperplasia, TIG1 methylation was either absent or low.59,66,94,110,111 Zhang et al94 further found that the methylation of TIG1 and RARβ was positively correlated. Therefore, it is possible that the methylation of the retinoid response gene TIG1 occurred in response to the methylation and inactivation of RARβ. Ellinger et al112 analyzed the diagnostic and prognostic possibilities of methylation analysis in cell-free serum DNA of patients with prostate cancer. They found that hypermethylation in TIG1 was more frequent in prostate cancer patients (10%) compared to BPH (0%) and healthy individuals (0%).59 The detection of hypermethylation in cell-free serum DNA may allow the specific diagnosis of prostate cancer.113
Tumor Cell Invasion and Metastasis Genes
Metastasis is an extremely complicated process that occurs through a series of sequential steps involving invasion, transport, adhesion at a distant site, and outgrowth into a secondary organ. Although metastases are the cause of 90% of human cancer deaths, little is known about the genetic and biochemical determinants of metastasis.
The methylated adenomatous polyposis coli (APC) gene causes familial adenomatous polyposis, which is an inherited disorder characterized by extensive colon polyps and the development of colorectal cancer in early adulthood. The APC complex is known to function as a gatekeeper in the cell, preventing the transcription of gene products that promote cell proliferation and survival rather than differentiation and apoptosis.114 Hypermethylation of APC implies silencing of this gatekeeper, making the cell vulnerable to further epigenetic and genetic changes and thus progression toward the development of invasive cancer. APC promoter methylation is common in various human tumors, especially in the colon. Most studies found a prevalence of 27% to 100% in prostate cancer tissue but only 5% to 6% in non cancerous tissue.17,19,20,22,27,28,45,48,55,66,115–118 Recent studies found that methylation in APC is associated with progression of prostate cancer.48,115,118 In two small cohorts of prostate cancer patients, a 3-fold statistically significantly increased hazard ratio (HR) for promoter methylation in APC has been reported among the patients who experienced prostate-specific antigen (PSA) recurrence, metastasis, or death.48,115 Richiardi et al118 found that hypermethylation in the promoter of the APC gene is involved in prostate cancer progression using large survival analysis.
Matrix metalloproteinases (MMPs) are proteolytic enzymes that degrade of the extracellular matrix and the basement membrane. High expressions of these enzymes have been associated with tumor growth, invasion, and tumor-induced angiogenesis.119 These pathways are controlled by the balance between the levels of the MMPs and tissue inhibitors of metalloproteinases (TIMPs).120 TIMP-2 and TIMP-3 are two of the frequently investigated members of this family because of their involvement in cancer progression and metastasis in a variety of human cancers.121–127 Pulukuri et al123 observed that 25 (60%) of 42 prostate tumors were methylated in TIMP-2 compared with 5 (16%) of 32 normal prostate samples. However, these results were not confirmed by a previous study.124 Ross et al124 found that TIMP-2 was expressed in a majority of prostate tumors and correlates with clinical stages. Contrary to the earlier study that indicated antitumor effects, TIMP-2 expression appears to have a tumor-promoting role in prostate cancer and warrants further investigation.124 High expression of TIMP-3 reduces metastasis, induces apoptosis, increases drug sensitivity, and in hibits tumor growth.125–127 A low expression by promoter methylation of TIMP-3 has been associated with poor outcomes.128 The promoter region of TIMP-3 was found to be methylated in 97% of prostate tumors.20 However, other studies reported low (6% and 0%) frequencies of TIMP-3 methylation.18,19 Two studies found TIMP-3 promoter methylation in 37% and 41% of urine sediments from prostate cancer patients.27,35 As a diagnostic marker in urine DNA, TIMP-3 may be limited by a persistent low frequency of methylation in normal controls.
Others tumor metastasis genes — Caveolin-1 (CAV1), E-cadherin (CDH1), H-cadherin (CDH13), EPHA7, and S100A2 — are often downregulated in prostate tumor tissues than in adjacent normal tissues due to methylation.17,19,27,28,39–42,91,129–135 Gene silencing of CAV1, CDH1, and CDH13 is associated with clinical features of prostate cancer.131,133,136,137 These data suggest that the methylation status of CAV1 and CDH1 not only is a potential biomarker for prostate cancer, but also may be a predictive marker of outcome.136 However, two studies reported that methylation of CDH1 promoter could not be detected in prostate cancer samples.19,41 S100A2 methylation was seen in 75% of cases of nonmalignant tissues and in 100% of cases of BPH.134
Conclusions
Although a few large-scale genome-wide analyses of epigenetic variations are currently ongoing, most published studies are small-scale with a retrospective design. Therefore, meta-analyses or large studies should be performed to obtain the complete extent and pattern of differential DNA methylation in the promoter region in the critical genes. Since epigenetic changes are involved in the carcinogenesis and progression of prostate cancer, information of these epigenetic changes may provide a clue for better diagnostic, prognostic, and predictive modalities than existing options. The ultimate goals of these epigenetic studies are to improve patient outcomes and enhance quality of life. A number of clinical trials and therapies are targeting methylated genes. Unlike DNA somatic mutations, DNA methylations are reversible. Thus, hypermethylated tumor-suppressor genes can be reactivated with drugs. Several demethylating agents such as 5-azacytidine (Vidaza) and 5-aza-2′-deoxycytidine (decitabine) have been approved as treatments for myelodysplastic syndrome (MDS) and leukemia.138–140 Some MDS patients treated with 5-azacytidine showed a significant survival benefit.141 However, a major limitation of these therapies is their nonspecific target approach, which may induce unintended side effects. Therefore, not only tumor suppressor genes but also silenced oncogenes by methylation can be reactivated. Future studies should focus on developing drugs that can target specific genes.
Footnotes
No significant relationship exists between the author and the companies/organizations whose products or services may be referenced in this article.
References
- 1.Crawford ED. Epidemiology of prostate cancer. Urology. 2003;62(6 suppl 1):3–12. doi: 10.1016/j.urology.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, et al. Cancer statistics, 2010. CA Cancer J Clin. 2010 Jul 7; Epub ahead of print. [Google Scholar]
- 3.Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16(4):168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- 4.Smiraglia DJ, Plass C. The study of aberrant methylation in cancer via restriction landmark genomic scanning. Oncogene. 2002;21(35):5414–5426. doi: 10.1038/sj.onc.1205608. [DOI] [PubMed] [Google Scholar]
- 5.Rush LJ, Dai Z, Smiraglia DJ, et al. Novel methylation targets in de novo acute myeloid leukemia with prevalence of chromosome 11 loci. Blood. 2001;97(10):3226–3233. doi: 10.1182/blood.v97.10.3226. [DOI] [PubMed] [Google Scholar]
- 6.Costello JF, Frühwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24(2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 7.Frühwald MC, O’Dorisio MS, Dai Z, et al. Aberrant promoter methylation of previously unidentified target genes is a common abnormality in medulloblastomas: implications for tumor biology and potential clinical utility. Oncogene. 2001;20(36):5033–5042. doi: 10.1038/sj.onc.1204613. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Herman JG, Graff JR, et al. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 9.Sulewska A, Niklinska W, Kozlowski M, et al. Detection of DNA methylation in eucaryotic cells. Folia Histochem Cytobiol. 2007;45(4):315–324. [PubMed] [Google Scholar]
- 10.Tanaka K, Okamoto A. Degradation of DNA by bisulfite treatment. Bioorg Med Chem Lett. 2007;17(7):1912–1915. doi: 10.1016/j.bmcl.2007.01.040. [DOI] [PubMed] [Google Scholar]
- 11.Park JY, Zheng W, Kim D, et al. Candidate tumor suppressor gene SLC5A8 is frequently down-regulated by promoter hypermethylation in prostate tumor. Cancer Detect Prev. 2007;31(5):359–365. doi: 10.1016/j.cdp.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg EC. How nucleotide excision repair protects against cancer. Nat Rev Cancer. 2001;1(1):22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 14.Mullaart E, Lohman PH, Berends F, et al. DNA damage metabolism and aging. Mutat Res. 1990;237(5–6):189–210. doi: 10.1016/0921-8734(90)90001-8. [DOI] [PubMed] [Google Scholar]
- 15.Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res. 2005;577(1–2):275–283. doi: 10.1016/j.mrfmmm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Park JY, Huang Y, Sellers TA. Single nucleotide polymorphisms in DNA repair genes and prostate cancer risk. Methods Mol Biol. 2009;471:361–385. doi: 10.1007/978-1-59745-416-2_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maruyama R, Toyooka S, Toyooka KO, et al. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2002;8(2):514–519. [PubMed] [Google Scholar]
- 18.Yamanaka M, Watanabe M, Yamada Y, et al. Altered methylation of multiple genes in carcinogenesis of the prostate. Int J Cancer. 2003;106(3):382–387. doi: 10.1002/ijc.11227. [DOI] [PubMed] [Google Scholar]
- 19.Yegnasubramanian S, Kowalski J, Gonzalgo ML, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64(6):1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 20.Jerónimo C, Henrique R, Hoque MO, et al. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10(24):8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. [DOI] [PubMed] [Google Scholar]
- 21.Konishi N, Nakamura M, Kishi M, et al. DNA hypermethylation status of multiple genes in prostate adenocarcinomas. Jpn J Cancer Res. 2002;93(7):767–773. doi: 10.1111/j.1349-7006.2002.tb01318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang GH, Lee S, Lee HJ, et al. Aberrant CpG island hypermethylation of multiple genes in prostate cancer and prostatic intraepithelial neoplasia. J Pathol. 2004;202(2):233–240. doi: 10.1002/path.1503. [DOI] [PubMed] [Google Scholar]
- 23.Li LC, Okino ST, Dahiya R. DNA methylation in prostate cancer. Biochim Biophys Acta. 2004;1704(2):87–102. doi: 10.1016/j.bbcan.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen TT, Nguyen CT, Gonzales FA, et al. Analysis of cyclin-dependent kinase inhibitor expression and methylation patterns in human prostate cancers. Prostate. 2000;43(3):233–242. doi: 10.1002/(sici)1097-0045(20000515)43:3<233::aid-pros10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 25.Jarrard DF, Bova GS, Ewing CM, et al. Deletional, mutational, and methylation analyses of CDKN2 (p16/MTS1) in primary and metastatic prostate cancer. Genes Chromosomes Cancer. 1997;19(2):90–96. [PubMed] [Google Scholar]
- 26.Gu K, Mes-Masson AM, Gauthier J, et al. Analysis of the p16 tumor suppressor gene in early-stage prostate cancer. Mol Carcinog. 1998;21(3):164–170. [PubMed] [Google Scholar]
- 27.Hoque MO, Topaloglu O, Begum S, et al. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J Clin Oncol. 2005;23(27):6569–6575. doi: 10.1200/JCO.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 28.Florl AR, Steinhoff C, Müller M, et al. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br J Cancer. 2004;91(5):985–994. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman JG, Merlo A, Mao L, et al. Inactivation of the CDKN2/p16/ MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55(20):4525–4530. [PubMed] [Google Scholar]
- 30.Nakamura M, Watanabe T, Klangby U, et al. p14ARF deletion and methylation in genetic pathways to glioblastomas. Brain Pathol. 2001;11(2):159–168. doi: 10.1111/j.1750-3639.2001.tb00388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin HH, Ke HL, Huang SP, et al. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol Oncol. 2009 Jan 30; doi: 10.1016/j.urolonc.2008.12.008. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 32.Chim CS, Chan WW, Kwong YL. Epigenetic dysregulation of the DAP kinase/p14/HDM2/p53/Apaf-1 apoptosis pathway in acute leukaemias. J Clin Pathol. 2008;61(7):844–847. doi: 10.1136/jcp.2007.047324. [DOI] [PubMed] [Google Scholar]
- 33.Calmon MF, Colombo J, Carvalho F, et al. Methylation profile of genes CDKN2A (p14 and p16), DAPK1, CDH1, and ADAM23 in head and neck cancer. Cancer Genet Cytogenet. 2007;173(1):31–37. doi: 10.1016/j.cancergencyto.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Konishi N, Nakamura M, Kishi M, et al. Heterogeneous methylation and deletion patterns of the INK4a/ARF locus within prostate carcinomas. Am J Pathol. 2002;160(4):1207–1214. doi: 10.1016/S0002-9440(10)62547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouprêt M, Hupertan V, Yates DR, et al. Molecular detection of localized prostate cancer using quantitative methylation-specific PCR on urinary cells obtained following prostate massage. Clin Cancer Res. 2007;13(6):1720–1725. doi: 10.1158/1078-0432.CCR-06-2467. [DOI] [PubMed] [Google Scholar]
- 36.Kuzmin I, Gillespie JW, Protopopov A, et al. The RASSF1A tumor suppressor gene is inactivated in prostate tumors and suppresses growth of prostate carcinoma cells. Cancer Res. 2002;62(12):3498–3502. [PubMed] [Google Scholar]
- 37.Liu L, Yoon JH, Dammann R, et al. Frequent hypermethylation of the RASSF1A gene in prostate cancer. Oncogene. 2002;21(44):6835–6840. doi: 10.1038/sj.onc.1205814. [DOI] [PubMed] [Google Scholar]
- 38.Kawamoto K, Okino ST, Place RF, et al. Epigenetic modifications of RASSF1A gene through chromatin remodeling in prostate cancer. Clin Cancer Res. 2007;13(9):2541–2548. doi: 10.1158/1078-0432.CCR-06-2225. [DOI] [PubMed] [Google Scholar]
- 39.Singal R, Ferdinand L, Reis IM, et al. Methylation of multiple genes in prostate cancer and the relationship with clinicopathological features of disease. Oncol Rep. 2004;12(3):631–637. [PubMed] [Google Scholar]
- 40.Woodson K, Hanson J, Tangrea J. A survey of gene-specific methylation in human prostate cancer among black and white men. Cancer Lett. 2004;205(2):181–188. doi: 10.1016/j.canlet.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 41.Woodson K, Hayes R, Wideroff L, et al. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55(3):199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 42.Padar A, Sathyanarayana UG, Suzuki M, et al. Inactivation of cyclin D2 gene in prostate cancers by aberrant promoter methylation. Clin Cancer Res. 2003;9(13):4730–4734. [PubMed] [Google Scholar]
- 43.Henrique R, Costa VL, Cerveira N, et al. Hypermethylation of Cyclin D2 is associated with loss of mRNA expression and tumor development in prostate cancer. J Mol Med. 2006;84(11):911–918. doi: 10.1007/s00109-006-0099-4. [DOI] [PubMed] [Google Scholar]
- 44.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121(11):2373–2380. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 45.Bastian PJ, Ellinger J, Wellmann A, et al. Diagnostic and prognostic information in prostate cancer with the help of a small set of hypermethylated gene loci. Clin Cancer Res. 2005;11(11):4097–4106. doi: 10.1158/1078-0432.CCR-04-1832. [DOI] [PubMed] [Google Scholar]
- 46.Kito H, Suzuki H, Ichikawa T, et al. Hypermethylation of the CD44 gene is associated with progression and metastasis of human prostate cancer. Prostate. 2001;49(2):110–115. doi: 10.1002/pros.1124. [DOI] [PubMed] [Google Scholar]
- 47.Gao X, Porter AT, Honn KV. Involvement of the multiple tumor suppressor genes and 12-lipoxygenase in human prostate cancer: therapeutic implications. Adv Exp Med Biol. 1997;407:41–53. doi: 10.1007/978-1-4899-1813-0_7. [DOI] [PubMed] [Google Scholar]
- 48.Rosenbaum E, Hoque MO, Cohen Y, et al. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin Cancer Res. 2005;11(23):8321–8325. doi: 10.1158/1078-0432.CCR-05-1183. [DOI] [PubMed] [Google Scholar]
- 49.Kim JW, Cheng Y, Liu W, et al. Genetic and epigenetic inactivation of LPL gene in human prostate cancer. Int J Cancer. 2009;124(3):734–738. doi: 10.1002/ijc.23972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen WY, Zeng X, Carter MG, et al. Heterozygous disruption of Hic1 predisposes mice to a gender-dependent spectrum of malignant tumors. Nat Genet. 2003;33(2):197–202. doi: 10.1038/ng1077. [DOI] [PubMed] [Google Scholar]
- 51.Chen W, Cooper TK, Zahnow CA, et al. Epigenetic and genetic loss of Hic1 function accentuates the role of p53 in tumorigenesis. Cancer Cell. 2004;6(4):387–398. doi: 10.1016/j.ccr.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 52.Weiss G, Cottrell S, Distler J, et al. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol. 2009;181(4):1678–1685. doi: 10.1016/j.juro.2008.11.120. [DOI] [PubMed] [Google Scholar]
- 53.Hampton T. New markers may help predict prostate cancer relapse risk. JAMA. 2006;295(19):2234–2238. doi: 10.1001/jama.295.19.2234. [DOI] [PubMed] [Google Scholar]
- 54.Vanaja DK, Ehrich M, Van den Boom D, et al. Hypermethylation of genes for diagnosis and risk stratification of prostate cancer. Cancer Invest. 2009;27(5):549–560. doi: 10.1080/07357900802620794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bastian PJ, Ellinger J, Heukamp LC, et al. Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing radical prostatectomy. Eur Urol. 2007;51(3):665–674. doi: 10.1016/j.eururo.2006.08.008. discussion 674. [DOI] [PubMed] [Google Scholar]
- 56.Lou W, Krill D, Dhir R, et al. Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res. 1999;59(10):2329–2331. [PubMed] [Google Scholar]
- 57.Nelson JB, Lee WH, Nguyen SH, et al. Methylation of the 5´ CpG island of the endothelin B receptor gene is common in human prostate cancer. Cancer Res. 1997;57(1):35–37. [PubMed] [Google Scholar]
- 58.Jerónimo C, Henrique R, Campos PF, et al. Endothelin B receptor gene hypermethylation in prostate adenocarcinoma. J Clin Pathol. 2003;56(1):52–55. doi: 10.1136/jcp.56.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ellinger J, Bastian PJ, Jurgan T, et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008;71(1):161–167. doi: 10.1016/j.urology.2007.09.056. [DOI] [PubMed] [Google Scholar]
- 60.Kekeeva TV, Popova OP, Shegaĭ PV, et al. Aberrant methylation of p16, HIC1, N33 and GSTP1 genes in tumor epitelium and tumor-associated stromal cells of prostate cancer [in Russian] Mol Biol (Mosk) 2007;41(1):79–85. [PubMed] [Google Scholar]
- 61.Murphy TM, Perry AS, Lawler M. The emergence of DNA methylation as a key modulator of aberrant cell death in prostate cancer. Endocr Relat Cancer. 2008;15(1):11–25. doi: 10.1677/ERC-07-0208. [DOI] [PubMed] [Google Scholar]
- 62.Park J, Brena RM, Gruidl M, et al. CpG island hypermethylation profiling of lung cancer using restriction landmark genomic scanning (RLGS) analysis. Cancer Biomark. 2005;1(2–3):193–200. doi: 10.3233/cbm-2005-12-307. [DOI] [PubMed] [Google Scholar]
- 63.Sørensen KD, Wild PJ, Mortezavi A, et al. Genetic and epigenetic SLC18A2 silencing in prostate cancer is an independent adverse predictor of biochemical recurrence after radical prostatectomy. Clin Cancer Res. 2009;15(4):1400–1410. doi: 10.1158/1078-0432.CCR-08-2268. [DOI] [PubMed] [Google Scholar]
- 64.Shivapurkar N, Toyooka S, Toyooka KO, et al. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int J Cancer. 2004;109(5):786–792. doi: 10.1002/ijc.20041. [DOI] [PubMed] [Google Scholar]
- 65.van Noesel MM, van Bezouw S, Salomons GS, et al. Tumor-specific down-regulation of the tumor necrosis factor-related apoptosis-inducing ligand decoy receptors DcR1 and DcR2 is associated with dense promoter hypermethylation. Cancer Res. 2002;62(7):2157–2161. [PubMed] [Google Scholar]
- 66.Cho NY, Kim BH, Choi M, et al. Hypermethylation of CpG island loci and hypomethylation of LINE-1 and Alu repeats in prostate adenocarcinoma and their relationship to clinicopathological features. J Pathol. 2007;211(3):269–277. doi: 10.1002/path.2106. [DOI] [PubMed] [Google Scholar]
- 67.Cheng Y, Kim JW, Liu W, et al. Genetic and epigenetic inactivation of TNFRSF10C in human prostate cancer. Prostate. 2009;69(3):327–335. doi: 10.1002/pros.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hornstein M, Hoffmann MJ, Alexa A, et al. Protein phosphatase and TRAIL receptor genes as new candidate tumor genes on chromosome 8p in prostate cancer. Cancer Genomics Proteomics. 2008;5(2):123–136. [PubMed] [Google Scholar]
- 69.Henderson BE, Ross RK, Pike MC, et al. Endogenous hormones as a major factor in human cancer. Cancer Res. 1982;42(8):3232–3239. [PubMed] [Google Scholar]
- 70.Henderson BE, Ross RK, Pike MC. Toward the primary prevention of cancer. Science. 1991;254(5035):1131–1138. doi: 10.1126/science.1957166. [DOI] [PubMed] [Google Scholar]
- 71.Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eder IE, Culig Z, Ramoner R, et al. Inhibition of LncaP prostate cancer cells by means of androgen receptor antisense oligonucleotides. Cancer Gene Ther. 2000;7(7):997–1007. doi: 10.1038/sj.cgt.7700202. [DOI] [PubMed] [Google Scholar]
- 73.Mitchell SH, Zhu W, Young CY. Resveratrol inhibits the expression and function of the androgen receptor in LNCaP prostate cancer cells. Cancer Res. 1999;59(23):5892–5895. [PubMed] [Google Scholar]
- 74.Tong Q, Zeng F, Lin C, et al. Growth inhibiting effects of antisense eukaryotic expression vector of proliferating cell nuclear antigen gene on human bladder cancer cells. Chin Med J (Engl) 2003;116(8):1203–1206. [PubMed] [Google Scholar]
- 75.Heisler LE, Evangelou A, Lew AM, et al. Androgen-dependent cell cycle arrest and apoptotic death in PC-3 prostatic cell cultures expressing a full-length human androgen receptor. Mol Cell Endocrinol. 1997;126(1):59–73. doi: 10.1016/s0303-7207(96)03970-6. [DOI] [PubMed] [Google Scholar]
- 76.Feldman BJ, Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 77.Grossmann ME, Huang H, Tindall DJ. Androgen receptor signaling in androgen-refractory prostate cancer. J Natl Cancer Inst. 2001;93(22):1687–1697. doi: 10.1093/jnci/93.22.1687. [DOI] [PubMed] [Google Scholar]
- 78.Kinoshita H, Shi Y, Sandefur C, et al. Methylation of the androgen receptor minimal promoter silences transcription in human prostate cancer. Cancer Res. 2000;60(13):3623–3630. [PubMed] [Google Scholar]
- 79.Sasaki M, Tanaka Y, Perinchery G, et al. Methylation and inactivation of estrogen, progesterone, and androgen receptors in prostate cancer. J Natl Cancer Inst. 2002;94(5):384–390. doi: 10.1093/jnci/94.5.384. [DOI] [PubMed] [Google Scholar]
- 80.Nakayama T, Watanabe M, Suzuki H, et al. Epigenetic regulation of androgen receptor gene expression in human prostate cancers. Lab Invest. 2000;80(12):1789–1796. doi: 10.1038/labinvest.3780190. [DOI] [PubMed] [Google Scholar]
- 81.Reibenwein J, Pils D, Horak P, et al. Promoter hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007;67(4):427–432. doi: 10.1002/pros.20533. [DOI] [PubMed] [Google Scholar]
- 82.Ho SM, Tang WY, Belmonte de Frausto J, et al. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66(11):5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hobisch A, Hittmair A, Daxenbichler G, et al. Metastatic lesions from prostate cancer do not express oestrogen and progesterone receptors. J Pathol. 1997;182(3):356–361. doi: 10.1002/(SICI)1096-9896(199707)182:3<356::AID-PATH863>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 84.Horvath LG, Henshall SM, Lee CS, et al. Frequent loss of estrogen receptor-beta expression in prostate cancer. Cancer Res. 2001;61(14):5331–5335. [PubMed] [Google Scholar]
- 85.Zhu X, Leav I, Leung YK, et al. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004;164(6):2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang X, Leung YK, Ho SM. AP-2 regulates the transcription of estrogen receptor (ER)-beta by acting through a methylation hotspot of the 0N promoter in prostate cancer cells. Oncogene. 2007;26(52):7346–7354. doi: 10.1038/sj.onc.1210537. [DOI] [PubMed] [Google Scholar]
- 87.Konishi N, Nakaoka S, Hiasa Y, et al. Immunohistochemical evaluation of estrogen receptor status in benign prostatic hypertrophy and in prostate carcinoma and the relationship to efficacy of endocrine therapy. Oncology. 1993;50(4):259–263. doi: 10.1159/000227191. [DOI] [PubMed] [Google Scholar]
- 88.Leav I, Lau KM, Adams JY, et al. Comparative studies of the estrogen receptors beta and alpha and the androgen receptor in normal human prostate glands, dysplasia, and in primary and metastatic carcinoma. Am J Pathol. 2001;159(1):79–92. doi: 10.1016/s0002-9440(10)61676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li LC, Chui R, Nakajima K, et al. Frequent methylation of estrogen receptor in prostate cancer: correlation with tumor progression. Cancer Res. 2000;60(3):702–706. [PubMed] [Google Scholar]
- 90.Nojima D, Li LC, Dharia A, et al. CpG hypermethylation of the promoter region inactivates the estrogen receptor-beta gene in patients with prostate carcinoma. Cancer. 2001;92(8):2076–2083. doi: 10.1002/1097-0142(20011015)92:8<2076::aid-cncr1548>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 91.Yao Q, He XS, Zhang JM, et al. Promotor hypermethylation of E-cadherin, p16 and estrogen receptor in prostate carcinoma [in Chinese] Zhonghua Nan Ke Xue. 2006;12(1):28–31. [PubMed] [Google Scholar]
- 92.Hayashi K, Yokozaki H, Naka K, et al. Overexpression of retinoic acid receptor beta induces growth arrest and apoptosis in oral cancer cell lines. Jpn J Cancer Res. 2001;92(1):42–50. doi: 10.1111/j.1349-7006.2001.tb01046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakayama T, Watanabe M, Yamanaka M, et al. The role of epigenetic modifications in retinoic acid receptor beta2 gene expression in human prostate cancers. Lab Invest. 2001;81(7):1049–1057. doi: 10.1038/labinvest.3780316. [DOI] [PubMed] [Google Scholar]
- 94.Zhang J, Liu L, Pfeifer GP. Methylation of the retinoid response gene TIG1 in prostate cancer correlates with methylation of the retinoic acid receptor beta gene. Oncogene. 2004;23(12):2241–2249. doi: 10.1038/sj.onc.1207328. [DOI] [PubMed] [Google Scholar]
- 95.Nelson CP, Kidd LC, Sauvageot J, et al. Protection against 2-hydroxyamino-1-methyl-6-phenylimidazo[4,5-b]pyridine cytotoxicity and DNA adduct formation in human prostate by glutathione S-transferase P1. Cancer Res. 2001;61(1):103–109. [PubMed] [Google Scholar]
- 96.Lee WH, Morton RA, Epstein JI, et al. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91(24):11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harden SV, Guo Z, Epstein JI, et al. Quantitative GSTP1 methylation clearly distinguishes benign prostatic tissue and limited prostate adenocarcinoma. J Urol. 2003;169(3):1138–1142. doi: 10.1097/01.ju.0000049627.90307.4d. [DOI] [PubMed] [Google Scholar]
- 98.Cairns P, Esteller M, Herman JG, et al. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7(9):2727–2730. [PubMed] [Google Scholar]
- 99.Lee WH, Isaacs WB, Bova GS, et al. CG island methylation changes near the GSTP1 gene in prostatic carcinoma cells detected using the polymerase chain reaction: a new prostate cancer biomarker. Cancer Epidemiol Biomarkers Prev. 1997;6(6):443–450. [PubMed] [Google Scholar]
- 100.Santourlidis S, Florl A, Ackermann R, et al. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39(3):166–174. doi: 10.1002/(sici)1097-0045(19990515)39:3<166::aid-pros4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 101.Goessl C, Krause H, Müller M, et al. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000;60(21):5941–5945. [PubMed] [Google Scholar]
- 102.Jerónimo C, Usadel H, Henrique R, et al. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer Inst. 2001;93(22):1747–1752. doi: 10.1093/jnci/93.22.1747. [DOI] [PubMed] [Google Scholar]
- 103.Gonzalgo ML, Pavlovich CP, Lee SM, et al. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9(7):2673–2677. [PubMed] [Google Scholar]
- 104.Jerónimo C, Varzim G, Henrique R, et al. I105V polymorphism and promoter methylation of the GSTP1 gene in prostate adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2002;11(5):445–450. [PubMed] [Google Scholar]
- 105.Köllermann J, Müller M, Goessl C, et al. Methylation-specific PCR for DNA-based detection of occult tumor cells in lymph nodes of prostate cancer patients. Eur Urol. 2003;44(5):533–538. doi: 10.1016/s0302-2838(03)00361-0. [DOI] [PubMed] [Google Scholar]
- 106.Goessl C, Müller M, Heicappell R, et al. DNA-based detection of prostate cancer in blood, urine, and ejaculates. Ann N Y Acad Sci. 2001;945:51–58. doi: 10.1111/j.1749-6632.2001.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 107.Suh CI, Shanafelt T, May DJ, et al. Comparison of telomerase activity and GSTP1 promoter methylation in ejaculate as potential screening tests for prostate cancer. Mol Cell Probes. 2000;14(4):211–217. doi: 10.1006/mcpr.2000.0307. [DOI] [PubMed] [Google Scholar]
- 108.Esteller M, Guo M, Moreno V, et al. Hypermethylation-associated Inactivation of the Cellular Retinol-Binding-Protein 1 Gene in Human Cancer. Cancer Res. 2002;62(20):5902–5905. [PubMed] [Google Scholar]
- 109.Jerónimo C, Henrique R, Oliveira J, et al. Aberrant cellular retinol binding protein 1 (CRBP1) gene expression and promoter methylation in prostate cancer. J Clin Pathol. 2004;57(8):872–876. doi: 10.1136/jcp.2003.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tokumaru Y, Sun DI, Nomoto S, et al. Re: Is TIG1 a new tumor suppressor in prostate cancer? J Natl Cancer Inst. 2003;95(12):919–920. doi: 10.1093/jnci/95.12.919. [DOI] [PubMed] [Google Scholar]
- 111.Topaloglu O, Hoque MO, Tokumaru Y, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10(7):2284–2288. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 112.Ellinger J, Haan K, Heukamp LC, et al. CpG island hypermethylation in cell-free serum DNA identifies patients with localized prostate cancer. Prostate. 2008;68(1):42–49. doi: 10.1002/pros.20651. [DOI] [PubMed] [Google Scholar]
- 113.Bastian PJ, Palapattu GS, Yegnasubramanian S, et al. CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. J Urol. 2008;179(2):529–534. doi: 10.1016/j.juro.2007.09.038. discussion 534–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer: a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 115.Henrique R, Ribeiro FR, Fonseca D, et al. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res. 2007;13(20):6122–6129. doi: 10.1158/1078-0432.CCR-07-1042. [DOI] [PubMed] [Google Scholar]
- 116.Tokumaru Y, Harden SV, Sun DI, et al. Optimal use of a panel of methylation markers with GSTP1 hypermethylation in the diagnosis of prostate adenocarcinoma. Clin Cancer Res. 2004;10(16):5518–5522. doi: 10.1158/1078-0432.CCR-04-0108. [DOI] [PubMed] [Google Scholar]
- 117.Enokida H, Shiina H, Urakami S, et al. Multigene methylation analysis for detection and staging of prostate cancer. Clin Cancer Res. 2005;11(18):6582–6588. doi: 10.1158/1078-0432.CCR-05-0658. [DOI] [PubMed] [Google Scholar]
- 118.Richiardi L, Fiano V, Vizzini L, et al. Promoter methylation in APC, RUNX3, and GSTP1 and mortality in prostate cancer patients. J Clin Oncol. 2009;27(19):3161–3168. doi: 10.1200/JCO.2008.18.2485. [DOI] [PubMed] [Google Scholar]
- 119.Gokaslan ZL, Chintala SK, York JE, et al. Expression and role of matrix metalloproteinases MMP-2 and MMP-9 in human spinal column tumors. Clin Exp Metastasis. 1998;16(8):721–728. doi: 10.1023/a:1006580728338. [DOI] [PubMed] [Google Scholar]
- 120.Gomez DE, Alonso DF, Yoshiji H, et al. Tissue inhibitors of metalloproteinases: structure, regulation and biological functions. Eur J Cell Biol. 1997;74(2):111–122. [PubMed] [Google Scholar]
- 121.Imren S, Kohn DB, Shimada H, et al. Overexpression of tissue inhibitor of metalloproteinases-2 retroviral-mediated gene transfer in vivo inhibits tumor growth and invasion. Cancer Res. 1996;56(13):2891–2895. [PubMed] [Google Scholar]
- 122.Mohanam S, Wang SW, Rayford A, et al. Expression of tissue inhibitors of metalloproteinases: negative regulators of human glioblastoma invasion in vivo. Clin Exp Metastasis. 1995;13(1):57–62. doi: 10.1007/BF00144019. [DOI] [PubMed] [Google Scholar]
- 123.Pulukuri SM, Patibandla S, Patel J, et al. Epigenetic inactivation of the tissue inhibitor of metalloproteinase-2 (TIMP-2) gene in human prostate tumors. Oncogene. 2007;26(36):5229–5237. doi: 10.1038/sj.onc.1210329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ross JS, Kaur P, Sheehan CE, et al. Prognostic significance of matrix metalloproteinase 2 and tissue inhibitor of metalloproteinase 2 expression in prostate cancer. Mod Pathol. 2003;16(3):198–205. doi: 10.1097/01.MP.0000056984.62360.6C. [DOI] [PubMed] [Google Scholar]
- 125.Han X, Zhang H, Jia M, et al. Expression of TIMP-3 gene by construction of a eukaryotic cell expression vector and its role in reduction of metastasis in a human breast cancer cell line. Cell Mol Immunol. 2004;1(4):308–310. [PubMed] [Google Scholar]
- 126.Deng X, Bhagat S, Dong Z, et al. Tissue inhibitor of metalloproteinase-3 induces apoptosis in prostate cancer cells and confers increased sensitivity to paclitaxel. Eur J Cancer. 2006;42(18):3267–3273. doi: 10.1016/j.ejca.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 127.Finan KM, Hodge G, Reynolds AM, et al. In vitro susceptibility to the pro-apoptotic effects of TIMP-3 gene delivery translates to greater in vivo efficacy versus gene delivery for TIMPs-1 or -2. Lung Cancer. 2006;53(3):273–284. doi: 10.1016/j.lungcan.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 128.Smith E, De Young NJ, Tian ZQ, et al. Methylation of TIMP3 in esophageal squamous cell carcinoma. World J Gastroenterol. 2008;14(2):203–210. doi: 10.3748/wjg.14.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bachmann N, Haeusler J, Luedeke M, et al. Expression changes of CAV1 and EZH2, located on 7q31 approximately q36, are rarely related to genomic alterations in primary prostate carcinoma. Cancer Genet Cytogenet. 2008;182(2):103–110. doi: 10.1016/j.cancergencyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 130.Cui J, Rohr LR, Swanson G, et al. Hypermethylation of the caveolin-1 gene promoter in prostate cancer. Prostate. 2001;46(3):249–256. doi: 10.1002/1097-0045(20010215)46:3<249::aid-pros1030>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 131.Li LC, Zhao H, Nakajima K, et al. Methylation of the E-cadherin gene promoter correlates with progression of prostate cancer. J Urol. 2001;166(2):705–709. [PubMed] [Google Scholar]
- 132.Alumkal JJ, Zhang Z, Humphreys EB, et al. Effect of DNA methylation on identification of aggressive prostate cancer. Urology. 2008;72(6):1234–1239. doi: 10.1016/j.urology.2007.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oudes AJ, Roach JC, Walashek LS, et al. Application of Affymetrix array and Massively Parallel Signature Sequencing for identification of genes involved in prostate cancer progression. BMC Cancer. 2005;5:86. doi: 10.1186/1471-2407-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rehman I, Cross SS, Catto JW, et al. Promoter hyper-methylation of calcium binding proteins S100A6 and S100A2 in human prostate cancer. Prostate. 2005;65(4):322–330. doi: 10.1002/pros.20302. [DOI] [PubMed] [Google Scholar]
- 135.Guan M, Xu C, Zhang F, et al. Aberrant methylation of EphA7 in human prostate cancer and its relation to clinicopathologic features. Int J Cancer. 2009;124(1):88–94. doi: 10.1002/ijc.23890. [DOI] [PubMed] [Google Scholar]
- 136.Karam JA, Lotan Y, Roehrborn CG, et al. Caveolin-1 overexpression is associated with aggressive prostate cancer recurrence. Prostate. 2007;67(6):614–622. doi: 10.1002/pros.20557. [DOI] [PubMed] [Google Scholar]
- 137.Lee SW. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat Med. 1996;2(7):776–782. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- 138.Mack GS. Epigenetic cancer therapy makes headway. J Natl Cancer Inst. 2006;98(20):1443–1444. doi: 10.1093/jnci/djj447. [DOI] [PubMed] [Google Scholar]
- 139.Müller CI, Rüter B, Koeffler HP, et al. DNA hypermethylation of myeloid cells, a novel therapeutic target in MDS and AML. Curr Pharm Biotechnol. 2006;7(5):315–321. doi: 10.2174/138920106778521523. [DOI] [PubMed] [Google Scholar]
- 140.Oki Y, Aoki E, Issa JP. Decitabine: bedside to bench. Crit Rev Oncol Hematol. 2007;61(2):140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 141.Müller A, Florek M. 5-Azacytidine/Azacitidine. Recent Results Cancer Res. 2010;184:159–170. doi: 10.1007/978-3-642-01222-8_11. [DOI] [PubMed] [Google Scholar]