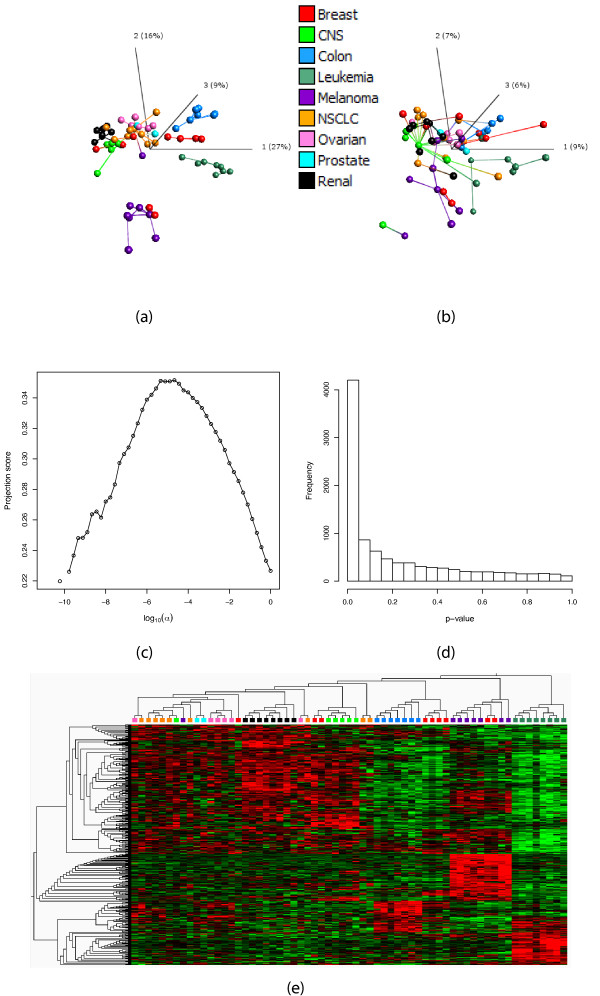
Figure 2.
Response-related filtering of the NCI-60 data. (a) The sample representation obtained by applying PCA to the most informative variable subset (482 variables) obtained by filtering with respect to the F-value for contrasting all nine cancer types. The different colors indicate different cancer types. (b) The sample representation obtained by applying PCA to the entire variable set (9,706 variables). (c) The projection score as a function of log10(α), where α is the p-value threshold used for inclusion. (d) The p-value distribution for all variables, indicating that there are truly significantly differentially expressed genes with respect to the contrast. (e) Heatmap of the most informative variable subset, that is, the one used to create the sample representation in (a). In panels (a) and (b), in order to obtain more easily interpretable plots, we joined the closest neighbors among the samples with line segments. The distance between two samples is defined by the Euclidean distance in the space spanned by all the remaining variables. The hierarchical clusterings in panel (e) are created using Euclidean distances and average linkage. The figures in (a), (b) and (e) were generated using Qlucore Omics Explorer 2.2 (Qlucore AB, Lund, Sweden).