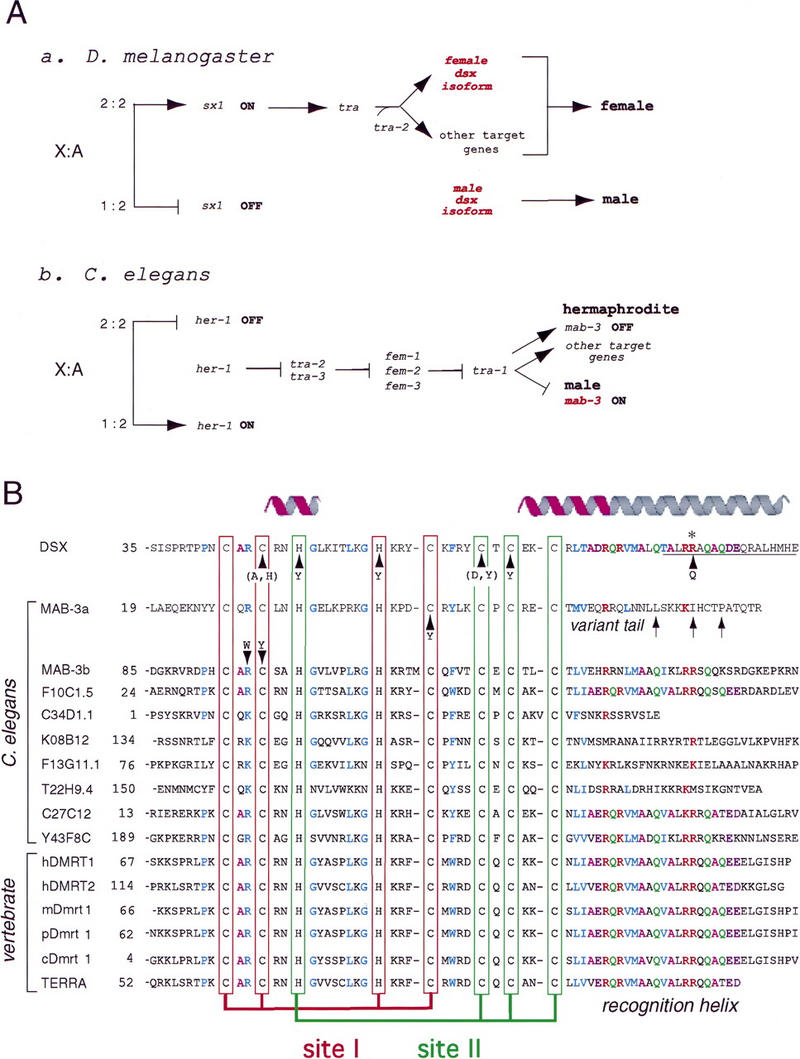
Figure 1.
(A) Genetic pathways of sex determination in D. melanogaster (a) and C. elegans (b) are initiated by X:autosome (A) ratio but otherwise different (Cline and Meyer 1996). (A,a) In the fly a high X:A ratio (2:2) activates sxl, which encodes a splicing factor. In turn, Sxl allows female-specific expression of the splicing factor tra, which together with tra-2, permits expression of female isoform DSXF. The pathway then ramifies. Intersex (ix), hermaphrodite (her), and fruitless (not shown) are other downstream elements. When the X:A ratio is low, male-specific isoform DSXM occurs by default; isoforms differ in carboxy-terminal domain (see Fig. 2A). (A,b) In nematode a high X:A ratio (2:2) is associated with absent her-1 expression, in turn enabling unrepressed expression of tra-2 and tra-3 (unrelated to Drosophila genes of similar nomenclature). Hermaphrodite-specific expression of tra-2 and tra-3 represses fem-1, fem-2, and fem-3. In their absence, tra-1 is expressed in hermaphrodite and turns off male-determinant mab-3 (encoding a DM transcription factor). TRA-1 has MAB-3 independent targets as pathway ramifies. When the X:A ratio is low (1:2), HER-1 (involved in cell–cell signaling) represses tra-2 and tra-3. Unrepressed expression of female genes represses tra-1 and therefore default expression of mab-3 directs male development and turns off hermaphroditic program. Nematode genes (b) that enumerate X:A ratio and control X dosage compensation (including xol-1 and sdc-2; Dawes et al. 1999) are omitted for clarity. (B) Alignment of metazoan DM sequence motifs. Cysteines and histidines that coordinate Zn2+ are aligned as two intertwined binding sites (boxes): site I (red) and site II (green). Conserved residues in zinc module are otherwise shown in blue; conserved residues in tail are color coded (R, red; Q, green; A, magenta; D/E, violet; L/V/T/M, blue). Stable α-helical elements are highlighted by magenta ribbons above DSX sequence. Nascent carboxy-terminal α-helix is indicated by gray extension; it is not meant to convey a continuous α-helix (see text). (DSX) DM domain in D. melanogaster (accession no. M25292). (MAB-3a and MAB-3b) The first and the second DM domains in C. elegans protein (accession no. Z99278). Other C. elegans DM sequences: F10C1.5, cosmid F10C1 (accession no. U49831); C34D1.1, cosmid C34D1 (accession no. Z78060); K08B12, cosmid K08B12 (accession no. U97001); F13G11, cosmid F13G11 (accession no. Z83317); T22H9.4: cosmid T22H9 (accession no. AF101315); C27C12 (accession no. Z69883); and Y43F8C (accession no. AL032637.1). Vertebrate DM sequences: DMRT1 and DMRT2, human homologs on the short arm of chromosome 9 (DMRT1: accession no. AF130728 and DMRT2: accession no. AF130729). Other homologs: mDmrt1 (murine, accession no. AF202778), pDmrt1 (porcine, accession no. AF216651), and cDmrt1 (chicken, accession no. AF123456). TERRA: DM domain of zebrafish terra (accession no. AF080622). Sequences shown include genes (such as TERRA; Meng et al. 1999) not known to be involved in sex determination. Arrows without parentheses indicate sites of point mutations in dsx or mab-3 associated with intersex development; substitutions in parenthesis indicate variants characterized only by biochemical assays (Erdman and Burtis 1993). Three additional putative DM genes of unknown function have recently been found in the genome of D. melanogaster (accession nos. AAF56919, AAF55843, and AAF48261; sequences not shown).