Figure 3.
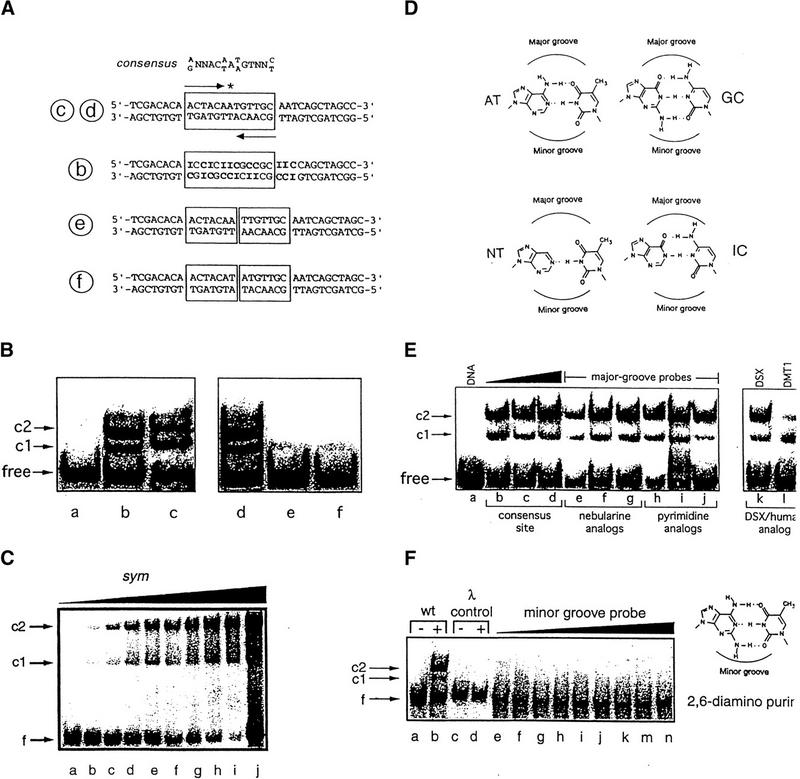
DNA-binding studies of DSX domain demonstrate minor groove binding mode. (A,top) Consensus sequence of DSX-binding sites, an imperfect palindrome containing an odd number of base pairs. (Second from top) DSX target site in fbe11–13. (Third from top) Analog containing multiple AT → IC substitutions (boldface type). (Bottom two sequences) Variants containing additional AT or TA base pairs in center. Labels b, c, d, e, f (circled) refer to lanes of gels in B. (B) 33P-detected GMSA at 4°C of DSX domain binding to sites defined in A. Bands c1 and c2 designate 1:1 and 2:1 protein–DNA complexes. Protein binds equally well to native and IC-containing sites. Insertion of central AT or TA base pairs reduces protein binding by about 100-fold. Protein concentration was 8 nm and DNA concentration was 0.5 nm. (C) Cooperative low-affinity binding of the DSX DM domain to a palindromic (sym) DNA site (5′-ACTACAATTGTTGCA-3′; central base pair in boldface type) using fluorescein (at 5′ of top strand) as probe. DNA concentration was 500 nm in lanes a–i, with respective protein concentrations 0, 250, 500, 600, 700, 800, 900, and 1000 nm. Lane j contains an equimolar solution in which concentrations of DNA and protein were each 5 μm; predominance of 2:1 complex and free DNA indicates cooperativity is retained. (D,left) Structures of AT and variant nebularine NT base pair. (D,right) Structures of GC and inosine IC base pair. Positions of major and minor grooves are indicated. (E,right gel) Major groove base modifications do not perturb binding of DSX domain to 15 base-pair DNA probe (5′-CACTACAATGTTGCA-3′ and complement): (lanes e–g) single nebularine substitutions at positions 5, 7, and 8 of upper strand and (lanes h–j) single 5-methylcytosine or uridine substituitions at positions 6, 9, and 11 of upper strand. (E,left gel) DMRT1 DM domain (25 nm) recognizes DSX target site with weaker apparent affinity; cooperative 2:1 binding is maintained. (F) 33P-detected GMSA of fbe analog in which the 8 AT bp in the DSX-binding site (boxed in Fig. 3A, sequence c,d) are substituted by 2,6-diaminopurine (structure at right). No binding is observed to λ operator site OL1 (lanes c,d) or to the diaminopurine fbe analog (lanes e–n) at successive protein concentrations of 0, 1, 2, 4, 8, 16, 32, 64, and 128 nm. DNA concentration was ∼1 nm. Protein concentrations were 8 nm for lanes b and d. Sequence of λ probe is 5′-TACCACTGGCGGTCATA-3′ and complement.