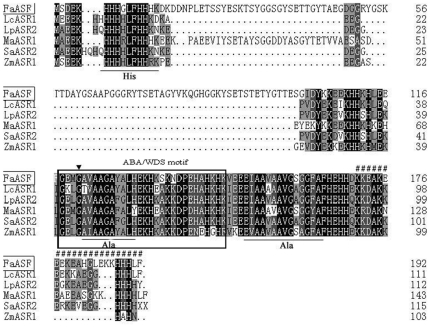
Figure 1. Amino acid sequence alignment of FaASR with other plant ASR proteins.
Strawberry FaASR was aligned with tomato LcASR1, LpASR2 and SaASR2, banana MpASR1 and maize ZmASR1. Conserved residues were shaded in black. Gray shading indicated similar residues in 3 out of 6 of the sequences. Gaps were introduced to optimize alignment. Multiple alignments was done by CLUSTALW and viewed with BOXSHADE program, and then manually edited. One Zn-binding His-rich region and two Ala-rich regions were underlined. The ABA/WDS motif was indicated with rectangle. One site for N-myristoylation and a putative nuclear targeting signal (KKEDKEEAEEASGKKHHH) were marked with ‘▾’ and ‘#’, respectively.