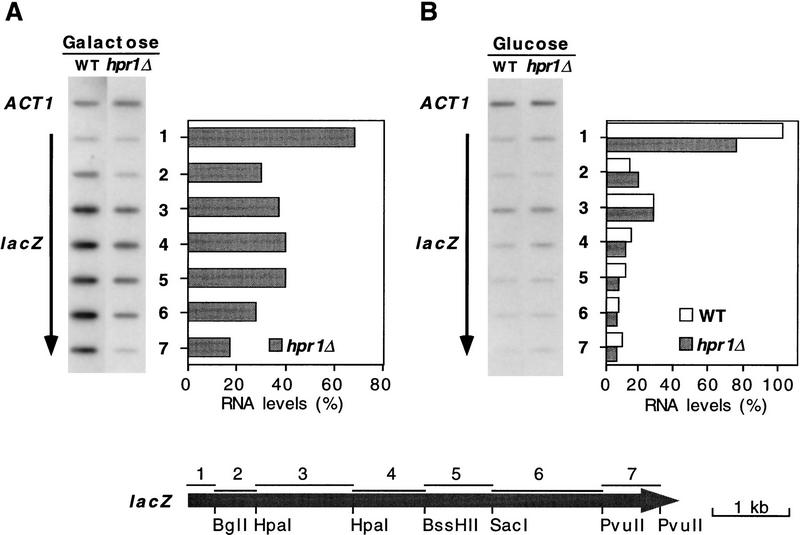
Figure 4.
Transcriptional run-on analysis in wild-type and hpr1Δ cells. Total RNA was isolated from wild-type and hpr1Δ cells transformed with single-copy p416GAL1–lacZ plasmid under induction (A) and repression (B) conditions. Two percent-galactose or 2% glucose was added to yeast cultures in glycerol–lactate synthetic medium at an OD600 of 0.05, 5 hr prior to the run-on analysis. The 0.6-kb internal ACT1 fragment and seven different DNA fragments (1–7) from the lacZ coding region were immobilized in hybond-N+ filters. The lacZ region covering each of the seven DNA fragments used is shown at the bottom. In all cases, the percentage of radiolabeled mRNA bound to each lacZ fragment was normalized with respect to their corresponding levels in galactose-grown wild-type cells, taken as 100% for each. The orientation of lacZ arrows indicates the direction of transcription. As negative control, we used DNA from Salmonella typhimurium (not shown).