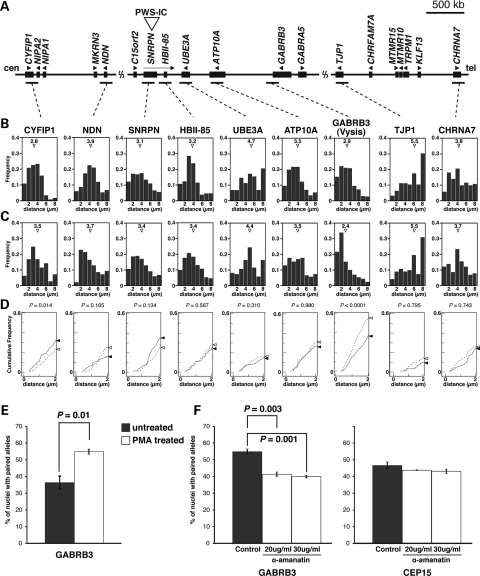
Figure 4.
Increased homologous pairing of GABRB3 alleles during neuronal differentiation is associated with active transcription. (A) Physical map of the imprinted gene cluster in human chromosome 15q11–q13. PWS-IC is the PWS imprinting center, which is essential for establishment of the paternal epigenetic state of the region. Genes or transcripts (filled boxes) are drawn approximately to scale. Transcriptional direction is indicated by arrowheads and arrows. The horizontal bars indicate BAC probes used in the pairing analyses. GABRB3(1) probe indicates the Vysis LSI GABRB3 Spectrum Orange probe. (B) Frequency distribution of distances between homologous alleles at multiple sites along 15q11–q13 for undifferentiated SH-SY5Y cell nuclei, as measured on each BAC probe. Mean distance, open triangle. (C) Frequency distribution of distances between homologous alleles at multiple sites along 15q11–q13 for PMA-treated differentiated SH-SY5Y cell nuclei. Mean distance, open triangle. A high proportion of PMA-treated differentiated SH-SY5Y cells displayed close homologous distances (≤2 µm) at GABRB3 upon neuronal differentiation, as shown by a leftward shift in the distribution. (D) Cumulative distribution of distances between homologous alleles at 0–2 µm. The solid line indicates the undifferentiated SH-SY5Y cells. The dotted line indicates the PMA-treated differentiated SH-SY5Y cells. The closed and open triangles indicate cumulative frequency at 2 µm, respectively. The statistical relevance was assessed by a comparison of the entire histogram of measurement distributions from (B) and (C) using two non-parametric tests, namely Mann–Whitney's U-test and Kolmogorov–Smirnov test. P-values from Mann–Whitney's U-test are as indicated. Sample sizes for each experiment ranged from 100 to 210. (E) Differences in homologous pairing in the undifferentiated SH-SY5Y cells (gray bars) and the PMA-treated differentiated SH-SY5Y cells (white bars), as determined by hybridization with Vysis LSI GABRB3 FISH probes. Pairing was scored as homologous FISH signals with distances ≤2 μm apart. The bars indicate the mean ± SEM of three replicate experiments, in each of which 100–150 nuclei were scored. (F) The PMA-treated differentiated SH-SY5Y cells treated for 4 h with the transcriptional inhibitor, α-amanitin, were hybridized with Vysis LSI GABRB3 (left panel) and CEP15 (right panel) FISH probes and signals measured. Transcriptional inhibition resulted in a significant reduction in the homologous pairing at GABRB3, but not at CEP15. Student's t-test.