Abstract
Background
Longitudinal mark-recapture studies of rodents in two sites in the Mbaracayú Biosphere Reserve in the Interior Atlantic Forest of eastern Paraguay have revealed a complex and intriguing pattern of hantaviruses harbored by rodents in this area. Full-length sequencing and phylogenetic analyses were conducted for several rodents from Akodon montensis and Oligoryzomys fornesi. The phylogenetic relationships of these viruses were analyzed in the context of hantaviruses in South America with published S- and M-segment sequences.
Findings
Phylogenetic analyses of hantaviruses identified in the Mbaracayú Biosphere Reserve in Paraguay revealed Jabora and Juquitiba viruses are harbored by Akodon montensis and Oligoryzomys fornesi, respectively. These analyses revealed that in general the constituents of the major subclade for the S- and M-segments differ for the South American hantaviruses. Further, the two major groups within subclade C for the M-segment reflect in general the lethality associated with the viruses within each group.
Conclusions
Phylogenetic studies of Jabora and Juquitiba viruses and other Paraguayan viruses in the context of American hantaviruses revealed reassortment and host-switching in the evolution of South American hantaviruses.
Keywords: Hantavirus; reassortment; host switching; zoonotic pathogens; ecology; emerging pathogens; phylogenetics; Akodon, Oligoryzomys
Findings
Numerous South American hantaviruses can cause hantavirus pulmonary syndrome (HPS) [1]. These viruses are harbored by rodents and studies suggest that each virus has coevolved with its unique rodent reservoir host, which allows viral persistence in the reservoir [2]. Our studies in the Mbaracayú Biosphere Reserve in the Interior Atlantic Forest of eastern Paraguay have revealed a complex and intriguing pattern of hantaviruses harbored by rodents in this area [3]. In Paraguay, Laguna Negra (LAN) virus in Calomys laucha was identified as the etiologic agent following an outbreak of HPS in western Paraguay [4].
We have conducted longitudinal mark-recapture studies of rodents in two sites in the Reserve (Jejui Mi and Horqueta Mi), and two outside (Rama III and Britez Cue) [3]. We have analyzed rodent population dynamics and hantavirus seroprevalence in this subtropical region [5] and reported on sympatry of hantaviruses in Akodon montensis and Oligoryzomys fornesi [3]. Full-length sequencing and phylogenetic analyses from several rodents from each species support these rodents as reservoirs of genotypes of Jabora virus (JABV) and Juquitiba virus (JUQV), respectively (Figure 1 and 2). JUQV is associated with HPS in cases in Brazil and in northeastern Argentina [6,7], while JABV-like viruses are not [8].
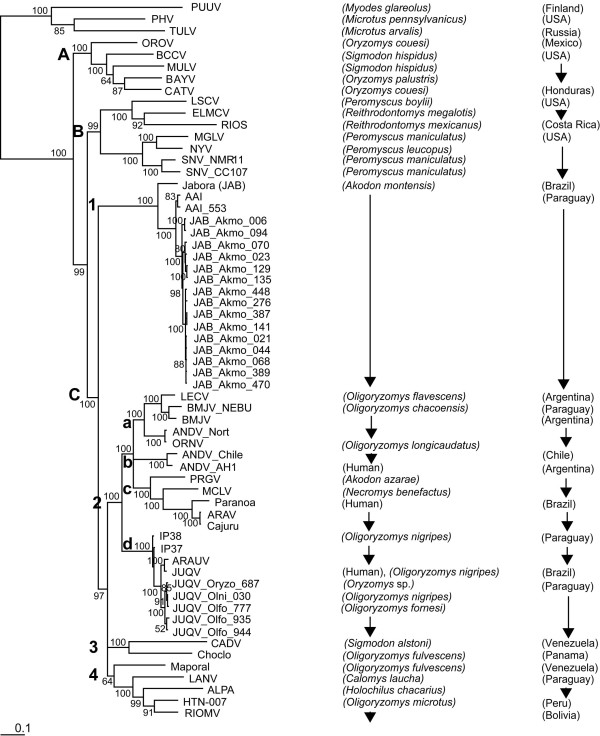
Figure 1.
Bayesian phylogenetic analysis of full length S-segment open reading frame nucleotide sequences of hantaviruses in Paraguay in the context of the Americas. Abbreviations: Akodon montensis is Akmo; Oligoryzomys fornesi is Olfo.; Clades are grouped A, B, C; Viral strains abbreviations include Puumula (PUU), Prospect Hill (PH), Tula (TUL), Oro (ORO), Black Creek Canal (BCC), Muleshoe (MUL), Bayou (BAY), Catacamas (CAT), Limestone Canyon (LSC), El Moro Canyon (ELMC), Rio Segunda (RIOS), MGLV, New York (NY), Sin number (SN), Jabora virus (JAB), Ape aime (AAI), Bermejo (BML), (ORN), Andes (AND), Pergamino (PERG), Itapua (IP), Araraquara (ARA), Juquitiba (JUQ), Caño Delgadito (CAD), Choclo, Laguna Negra (LAN), Rio Mamoré (RIOM), Maporal.
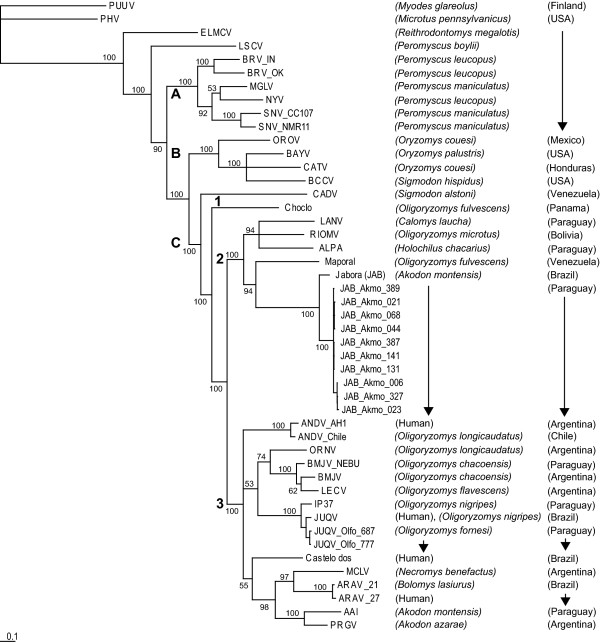
Figure 2.
Bayesian phylogenetic analysis of full length M-segment open reading frame nucleotide sequences of hantaviruses in Paraguay in the context of the Americas. Abbreviations: Akodon montensisis Akmo; Oligoryzomys fornesiis Olfo.; Clades are grouped A, B, C; Viral strains abbreviations include Puumula (PUU), Prospect Hill (PH), Tula (TUL), Oro (ORO), Black Creek Canal (BCC), Muleshoe (MUL), Bayou (BAY), Catacamas (CAT), Limestone Canyon (LSC), El Moro Canyon (ELMC), Rio Segunda (RIOS), MGLV, New York (NY), Sin number (SN), Jabora virus (JAB), Ape aime (AAI), Bermejo (BML), (ORN), Andes (AND), Pergamino (PERG), Itapua (IP), Araraquara (ARA), Juquitiba (JUQ), Caño Delgadito (CAD), Choclo, Laguna Negra (LAN), Rio Mamoré (RIOM), Maporal.
We have made Baysian phylogenetic analyses of these and published S and M sequences that have at least one Kb of sequence from American hantaviruses (Figure 1 and 2). In Figure 1 and 2, for the JABV and JUQV identified in the Reserve, the genotype is indicated as the viral strain followed by the rodent reservoir and identification number. The phylogenetic relationships of subclade C reveal several features in the evolution of hantaviruses in Paraguay and in South America when comparisons are made between S- and M-segments. First, these phylogenetic analyses revealed that in general the constituents of the subclade C for the S- and M-segments differ for the South American hantaviruses. For the S-segment, the phylogenetic tree for subclade C shows four subgroups: C1-JAB strains (Brazil, Paraguay), and a strain (AAI) from Itapúa, Paraguay; C2- Andes (AND), Araraquara (ARA), and Juquitiba (JUQ) viruses (Argentina, Brazil, Chile, Paraguay); C3- Caño Delgadito (CAD), and Choclo viruses (Venezula, Panama); and, C4- LAN, Rio Mamoré (RIOM), Maporal viruses (Bolivia, Paraguay, Peru, Venezula). All the subclade C groups show strong bootstrap values except for C4, which is due to the sequence of Maporal virus. For the M-segment, the phylogenetic tree for subclade C shows three subgroups: C1- Choclo; C2-LAN, RIOM, JAB viruses; and, C3- AND, ARA, JUQ viruses. Notably, the M-segment analyses improved the low bootstrap value for the group with Maporal virus through creation of an independent subgroup (compare M-segment C2 and S-segment C4).
Secondly, we noted two major groups within subclade C for the M-segment that reflect in general the lethality associated with the viruses within each group. Most of the viruses in subclade C2 show a mortality in HPS cases of ~5-10%. In contrast, many of the viruses in subclade C3 show a very high mortality of 40-50%. The S-segment phylogenetic tree further subdivides the M-segment C2 subclade into the JABV group (S-segment C1) and the LANV/RIOM group (S-segment C4). At present, we have no information on whether viruses of the JABV group cause HPS. The amino acid homologies of representatives of these viruses also break into two major groups based on low or high severity of HPS (Table 1).
Table 1.
Amino acid sequence similarity of S and M segment among hantaviruses identified in Paraguay and nearby countries
| LAN | RIO M | ALPA | JAB | JAB Akmo 006 | AAI | AND | BMJ-NEB | IP37 | JUQV Olfo 777 | JUQ | PRG | ARA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LAN | - | 96 | 94 | 92 | 90 | 94 | 94 | 94 | 94 | 94 | 94 | 94 | NA |
| RIOM | 93 | -- | 94 | 92 | 90 | 92 | 94 | 94 | 94 | 94 | 94 | 93 | NA |
| ALPA | 92 | 96 | -- | 94 | 94 | 95 | 94 | 94 | 96 | 96 | 96 | 95 | NA |
| JAB | 85 | 89 | 88 | -- | 97 | 94 | 93 | 93 | 94 | 94 | 95 | 95 | NA |
| JAB Akmo 006 | 86 | 88 | 88 | 99 | -- | 94 | 93 | 93 | 94 | 94 | 95 | 95 | NA |
| AAI | 88 | 90 | 90 | 99 | 99 | - | 95 | 95 | 96 | 96 | 96 | 96 | NA |
| AND | 90 | 90 | 89 | 86 | 86 | 88 | - | 98 | 98 | 96 | 96 | 95 | NA |
| BMJ-NEB | 89 | 90 | 88 | 86 | 85 | 88 | 98 | - | 98 | 97 | 98 | 98 | NA |
| IP37 | 90 | 90 | 89 | 86 | 86 | 88 | 96 | 95 | - | 99 | 99 | 98 | NA |
| JUQ Olfo 777 | 89 | 88 | 87 | 86 | 86 | 87 | 96 | 94 | 100 | - | 99 | 99 | NA |
| JUQ | 90 | 90 | 89 | 86 | 85 | 88 | 96 | 95 | 100 | 100 | - | 97 | NA |
| PRG | 90 | 90 | 89 | 85 | 84 | 87 | 96 | 95 | 93 | 92 | 93 | - | NA |
| ARA | 91 | 91 | 90 | 90 | 89 | 90 | 96 | 94 | 94 | 94 | 94 | 96 | - |
Above -: M segment amino acid sequence similarity
Below -: S segment amino acid sequence similarity
Our analyses further revealed a reassortment of AAIV (harbored by A. montensis), identified in Itapúa, Paraguay [9], that falls in subclade C3 in the M-segment. AAIV shows a strong relationship with Pergamino virus (PRGV), originally identified in Argentina in A. azarae. However, for the S-segment, AAIV shows a strong relationship with JABV. In agreement with in vitro published reassortant studies of ANDV [10], the AAIV genotype was a reassortment of the S-segment of the JABV-like viral genotypes and the M-segment of the AND-like viral genotypes. The direction of the reassortment would suggest spillover of the AND-like viral genotype into an A. montensis, which would have necessarily harbored a JABV at that time. Even more intriguing is the grouping of JABV in the S- and M-Segment analyses. The JABV strain clearly shows a strong relationship in the M-segment C-2 subclade with the LAN and RIOM viral strains. However, the S-segment analyses reveal that the JAB and AAI are more closely related, and remarkably, these strains do not cluster with any of the other subclades, C2-4.
Finally, the phylogenetic trees do not support strong associations of any host phyletic groups within any subclade. The lack of association with the rodent groups argues that unlike hantaviruses in Europe and Asia [2], hantaviruses do not show the level of coevolution with their hosts in South America and hence these viruses have great potential for host switching and adaptation. Certainly, the recent radiation of the Sigmodontinae in South America [11,12] reflects a more recent introduction of hantaviruses into South America. Molecular clock analyses suggest that hantaviruses were introduced approximately 800 years ago [13].
In summary, future studies that integrate large scale phylogeographic mapping coupled with local molecular phylogenetic analyses of rodent-hantavirus relationships in the Americas have great potential to address important questions in the ecology of zoonotic pathogens such as the molecular events that lead to transfer and adaptation to a new host. In South America, events that lead to transmission, host switching, recombination, reassortment and post-transfer adaptation have not been addressed. These questions are critical to interpretation of ecological trends in the emergence and spread of zoonotic diseases, causes of outbreaks, and importantly, guidelines for control and prevention of disease.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
YKC participated in the cloning, sequencing, sequence alignment and phylogenetic analyses. YKC, RDO and CBJ participated in the design of the study. YKO, RDO and CBJ conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yong-Kyu Chu, Email: y0chu001@louisville.edu.
Robert D Owen, Email: rowen@tigo.com.py.
Colleen B Jonsson, Email: cbjons01@louisville.edu.
Acknowledgements
We thank Robert J. Baker and Heath Garner of the Museum of Texas Tech University for approving and facilitating loans of rodent tissues; the Secretaría del Ambiente (Paraguay) for permits to collect and export rodents and tissues; and the field crew, led by Ismael Mora, for dedication to their work, regardless of circumstances. This work was supported by a grant from the Fogarty International Center 1 R01 TW006986-01 to CBJ under the NIH-NSF Ecology of Infectious Diseases initiative.
References
- Jonsson CB, Figueiredo LT, Vapalahti O. A global perspective on hantavirus ecology, epidemiology, and disease. Clin Microbiol Rev. 2010;23(2):412–441. doi: 10.1128/CMR.00062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plyusnin A, Morzunov SP. Virus evolution and genetic diversity of hantaviruses and their rodent hosts. Curr Top Microbiol Immunol. 2001;256:47–75. doi: 10.1007/978-3-642-56753-7_4. [DOI] [PubMed] [Google Scholar]
- Chu YK, Goodin D, Owen RD, Koch D, Jonsson CB. Sympatry of 2 hantavirus strains, paraguay, 2003-2007. Emerg Infect Dis. 2009;15(12):1977–1980. doi: 10.3201/eid1512.090338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Peters CJ, Nichol ST. Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238(1):115–127. doi: 10.1006/viro.1997.8840. [DOI] [PubMed] [Google Scholar]
- Owen RD, Goodin DG, Koch DE, Chu Y, Jonsson CB. Spatiotemporal variation in Akodon montensis (Cricetidae: Sigmodontinae) and hantaviral seroprevalence in a subtropical forest ecosystem. J Mammalogy. 2010;91:467–481. doi: 10.1644/09-MAMM-A-152.1. [DOI] [Google Scholar]
- Johnson AM, de Souza LT, Ferreira IB, Pereira LE, Ksiazek TG, Rollin PE, Peters CJ, Nichol ST. Genetic investigation of novel hantaviruses causing fatal HPS in Brazil. J Med Virol. 1999;59(4):527–535. doi: 10.1002/(SICI)1096-9071(199912)59:4<527::AID-JMV17>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Padula P, Martinez VP, Bellomo C, Maidana S, San Juan J, Tagliaferri P, Bargardi S, Vazquez C, Colucci N, Estevez J. et al. Pathogenic hantaviruses, northeastern Argentina and eastern Paraguay. Emerg Infect Dis. 2007;13(8):1211–1214. doi: 10.3201/eid1308.061090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raboni SM, Hoffmann FG, Oliveira RC, Teixeira BR, Bonvicino CR, Stella V, Carstensen S, Bordignon J, D'Andrea PS, Lemos ER, Phylogenetic characterization of hantaviruses from wild rodents and HPS cases in the state of Parana (Southern Brazil) J Gen Virol. 2009. [DOI] [PubMed]
- Chu YK, Milligan B, Owen RD, Goodin DG, Jonsson CB. Phylogenetic and geographical relationships of hantavirus strains in eastern and western Paraguay. Am J Trop Med Hyg. 2006;75(6):1127–1134. [PMC free article] [PubMed] [Google Scholar]
- McElroy AK, Smith JM, Hooper JW, Schmaljohn CS. Andes virus M genome segment is not sufficient to confer the virulence associated with Andes virus in Syrian hamsters. Virology. 2004;326(1):130–139. doi: 10.1016/j.virol.2004.05.018. [DOI] [PubMed] [Google Scholar]
- D'Elia G, Luna L, Gonzalez EM, Patterson BD. On the Sigmodontinae radiation (Rodentia, Cricetidae): an appraisal of the phylogenetic position of Rhagomys. Mol Phylogenet Evol. 2006;38(2):558–564. doi: 10.1016/j.ympev.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Miranda GB, Oliveira LF, Andrades-Miranda J, Langguth A, Callegari-Jacques SM, Mattevi MS. Phylogenetic and phylogeographic patterns in sigmodontine rodents of the genus oligoryzomys. J Hered. 2009;100(3):309–321. doi: 10.1093/jhered/esn099. [DOI] [PubMed] [Google Scholar]
- Ramsden C, Melo FL, Figueiredo LM, Holmes EC, Zanotto PM. High rates of molecular evolution in hantaviruses. Mol Biol Evol. 2008;25(7):1488–1492. doi: 10.1093/molbev/msn093. [DOI] [PubMed] [Google Scholar]