Figure 2.
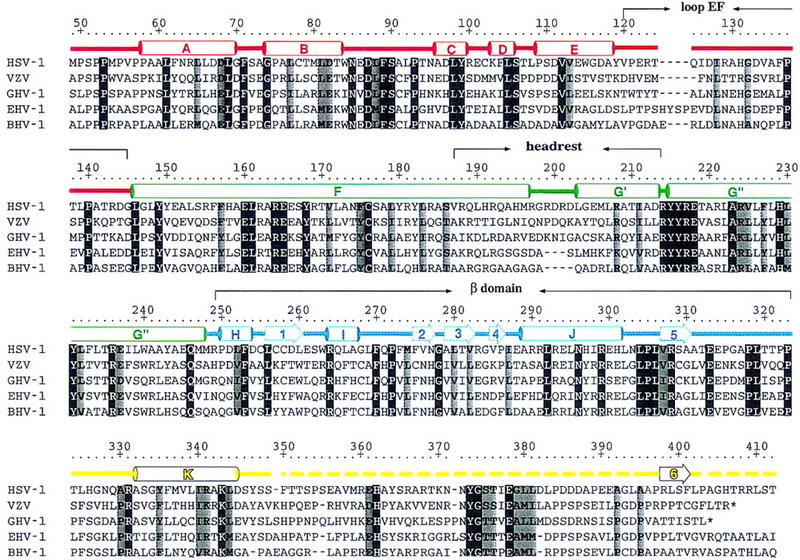
Sequence alignment of the conserved VP16-core region from HSV-1, VZV, GHV-1, EHV-1, and BHV-1 and alignment with structural elements of the VP16-core protein. The HSV-1 residue numbering is shown above the sequence alignment. Amino acids highlighted are either invariant (white against black) among the five proteins or similar (shaded) as defined by the following groupings: V, L, I, and M; F, Y, and W; K and R; E and D; Q and N; S and T; and A, G, and P. The color coding is as described in the text; α helices are labeled A–K (helix αG is subdivided into helices αG′ and αG′′ owing to a discontinuity in this helix); β strands are labeled 1–6; turns and loops (L) are referred to by their flanking secondary structures; broken lines indicate disordered regions. Polar interactions among conserved residues that are seen to exert their effect on the structure of VP16 as a whole include ion pairings (R64–D68, D97–R162, E160–H131, R164–E237, E165–K343, R214–E218, and R308–E313), polar side-chain– side-chain interactions (D87–S90, Y149–Q246, Y99–E165, S106–D111, Y168–H229, Y231–K343), polar side-chain–main-chain interactions (N85–H326, N85–A330, S106–L108, S186–Y182, Y215–F104, Y216–S106, R237–L302, R264–S90, N296–V307, R299–P305, R299–L269), and water-mediated polar interactions (S106-H2O–S348, E160-H2O–E237, Y182-H2O–R214, Y335-H2O–A330).
