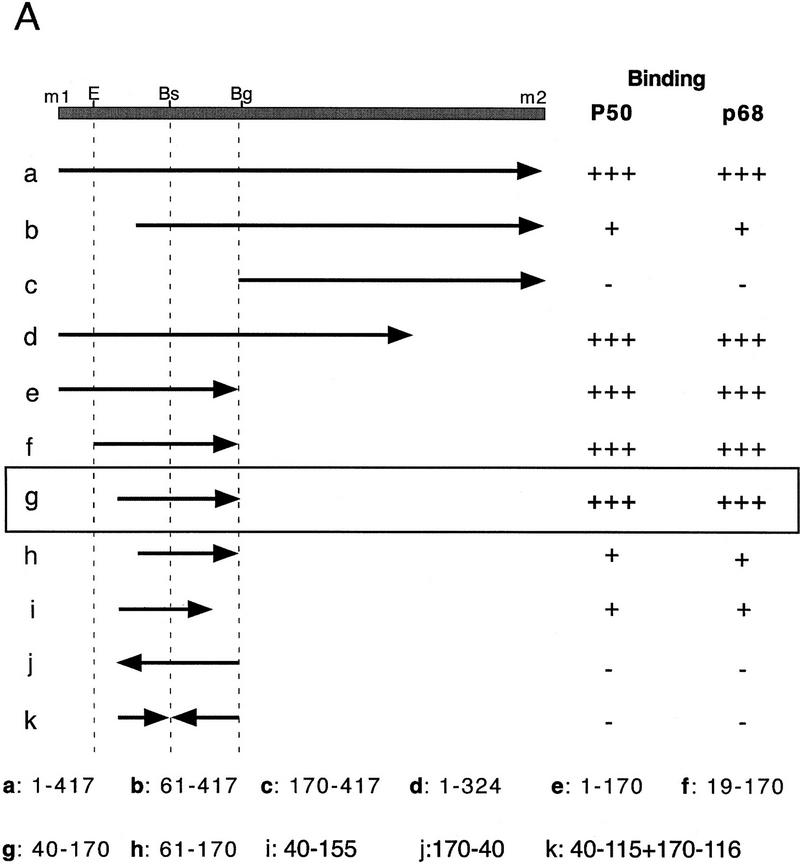
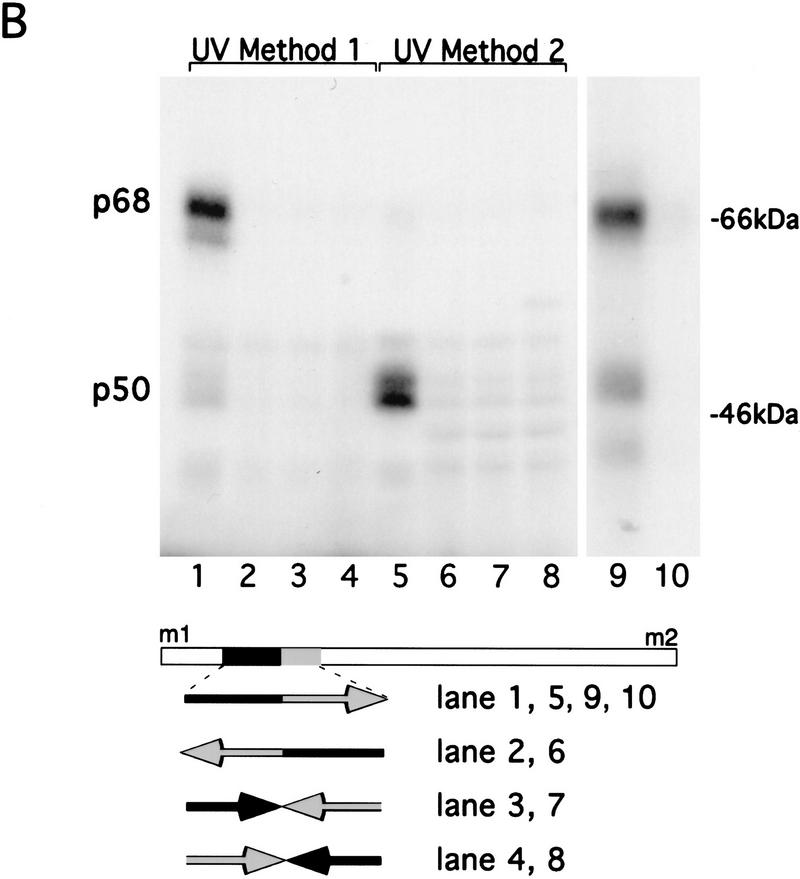
Figure 3.
Mapping specific protein/RNA interactions in the m1–m2 fragment of osk mRNA that is essential for its translation. (A) The 5′ end of osk mRNA, including the first and second start codons, m1 and m2, respectively, is diagrammed schematically. A partial restriction map of the region, showing sites used for subcloning is shown at the top. (E) EcoRI; (Bs) BstEII; (Bg) BglII. Transcripts used as probes for the UV cross-linking assay are identified by lowercase letters. The coordinates of the subfragments are indicated at the bottom. Position 1 is the first nucleotide of m1, position 417 is the last nucleotide of m2. The entire m1–m2 region (a) or subfragments of the region, created with available restriction sites or by PCR were labeled radioactively and incubated with oocyte extract. Proteins cross-linked to labeled RNA were separated by SDS-PAGE and visualized by autoradiography. The signal intensity obtained with the full-length m1–m2 probe was used as a standard and is indicated by (+++). Significantly reduced but detectable binding, and complete loss of binding are indicated (+) and (−), respectively. (B) UV cross-linking assay revealing the specific interaction of p50 and p68 with RNA fragment g. RNA probes (bottom) were incubated with oocyte extract and cross-linked by method 1 (lanes 1–4 and 9,10) or method 2 (lanes 5–8). (Lane 10) p68 complex with RNA fragment g was incubated with anti-Bruno antiserum and subjected to immunoprecipitation. The minimal region recognized (fragment g) was determined as shown in A. Probes used in each lane are indicated below. Arrows pointing rightward indicate RNA in sense orientation; arrows pointing leftward indicate RNA in antisense orientation.